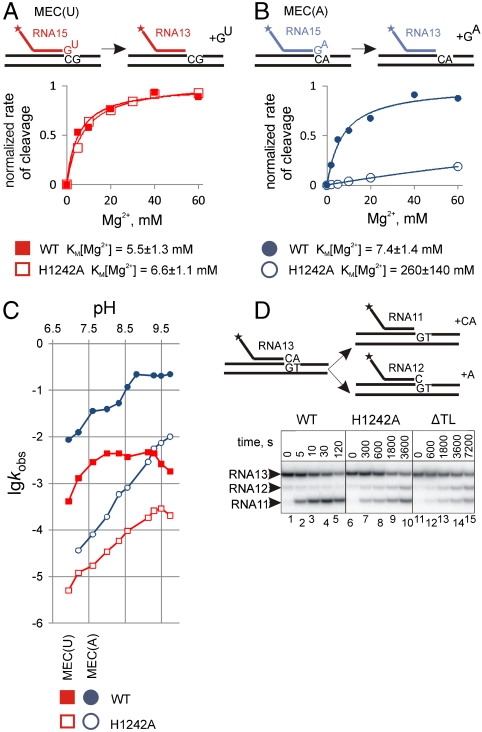
Fig. 2.
H1242 of the TL directly participates in phosphodiester bond cleavage. (A and B) Mg2+ dependences of the hydrolysis of the second phosphodiester bond in MEC(U) and MEC(A), respectively, by WT and H1242A RNAPs. Schematics above the plots show the reactions; the asterisk indicates that RNA is labeled at the 5′ end. The fits of data to the Michaelis–Menten equation are shown as solid lines. The kcat (reaction rate in saturating Mg2+) of the reactions were taken as 1 for clarity. KM[Mg2+] values are shown below the plots. (C) pH profiles of second phosphodiester bond hydrolysis in MEC(U) (Red) and MEC(A) (Blue) complexes (colors correspond to reactions shown in A and B) by WT (Solid Squares and Circles, respectively) and H1242A (Empty Squares and Circles, respectively) RNAPs. (D) The first and the second phosphodiester bond hydrolysis in the elongation complex with the correctly paired 3′ end AMP of the RNA (schematically shown above the gel; EC1 in Fig. S1) by WT, H1242A, and ΔTL RNAPs.