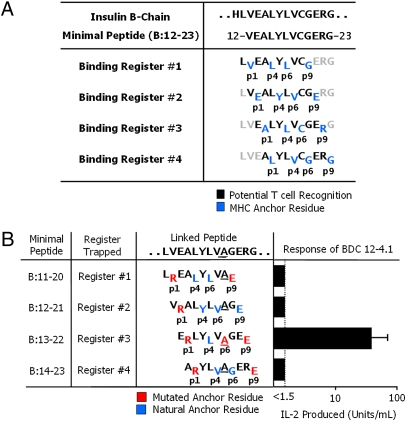
Fig. 1.
The BDC 12-4.1 T cell responds to register 3 of the insulin B:12–23 peptide. (A) Four possible IAg7-binding registers for the mouse insulin B-chain peptide, B:12–23. Anchor residues are shown in blue, and black residues show the amino acids that potentially contact the T-cell antigen receptor. (B) Minimal 10-mer linked peptide versions of the insulin B:12–23(19A) epitope. Each minimal peptide was trapped in a single binding register by a p1Arg and a p9Glu. The substitution of Ala for Cys at B:19 of the peptide is underlined. Red amino acids are the substituted anchor residues. Blue amino acids are the naturally occurring anchor residues. Black amino acids are the possible T-cell epitope residues. Also shown is the stimulation of the BDC 12-4.1 T-cell hybridoma with each of the individual IAg7-linked peptide complexes expressed on the surface of ICAM/B7+ insect cells, assessed by the production of IL-2. Results are the average ± SEM of triplicate wells from five independent experiments.