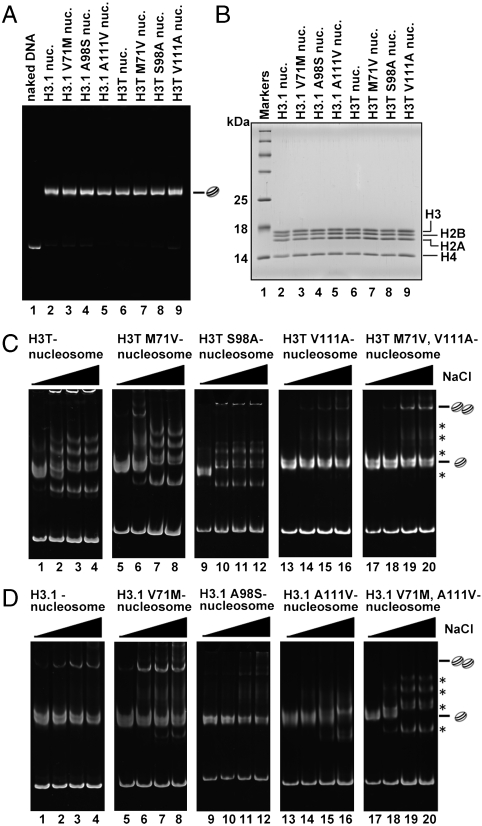
Fig. 5.
Mutational analysis of H3T and H3.1 nucleosomes. (A) Nucleosomes containing H3T and H3.1 mutants were purified using a Prepcell apparatus and were analyzed by nondenaturing 6% PAGE with ethidium bromide staining. (B) Histone compositions of the purified nucleosomes containing H3T and H3.1 mutants were analyzed by 18% SDS-PAGE with Coomassie brilliant blue staining. (C and D) Salt titration. Nucleosomes were incubated in the presence of 0.4 M (lanes 1, 5, 9, 13, and 17), 0.6 M (lanes 2, 6, 10, 14, and 18), 0.7 M (lanes 3, 7, 11, 15, and 19), and 0.8 M NaCl (lanes 4, 8, 12, 16, and 20) at 42 °C for 2 h. The samples were analyzed by nondenaturing 6% PAGE with ethidium bromide staining. Bands corresponding to nucleosome monomers and nucleosome–nucleosome aggregates are indicated. Asterisks represent bands corresponding to nonnucleosomal DNA-histone complexes. (C) The stability of H3T mutants. Lanes: 1–4, H3T; 5–8, H3T-M71V; 9–12, H3T-S98A; 13–16, H3T-V111A; and 17–20, H3T-M71V/V111A nucleosomes. (D) The stability of H3.1 mutants. Lanes: 1–4, H3.1; 5–8, H3.1-V71M; 9–12, H3.1-A98S; 13–16, H3.1-A111V; and 17–20, H3.1-V71M/A111V.