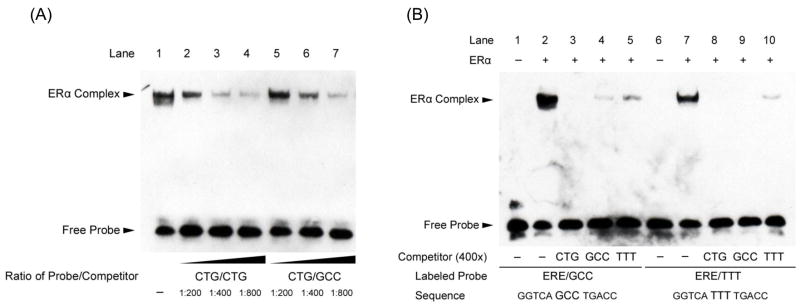
Fig. 2.
Binding affinity for the ERE with a CTG spacer sequence is greater than for EREs with GCC or TTT spacer sequences. (A) ERα binding to the consensus ERE sequence (GGTCACTGTGACC) is shown in lane 1. Competition using serial dilutions of the same unlabeled DNA sequence (lanes 2, 3, and 4) or unlabeled sequences with the variant spacer sequence GCC (lanes 5, 6, and 7) is shown. The unlabeled competitor with ERE spacer CTG demonstrated higher affinity binding to ERα than did the GCC-spaced ERE sequence. (B) Complementary EMSA experiments confirm that ERα preferentially binds EREs with a CTG tri-nucleotide spacer sequence. ERα binding to two consensus EREs with variant tri-nucleotide spacers is shown: GGTCAGCCTGACC (lanes 2–5) and GGTCATTTTGACC (lanes 7–10). Competition using 400 fold excess of the indicated unlabeled ERE sequences is also shown and reveals that relative efficiency of competition follows the order CTG (lanes 3 and 8) > GCC (lanes 4 and 9) > TTT (lanes 5 and 10). Shown are representative experiments performed at least three times.