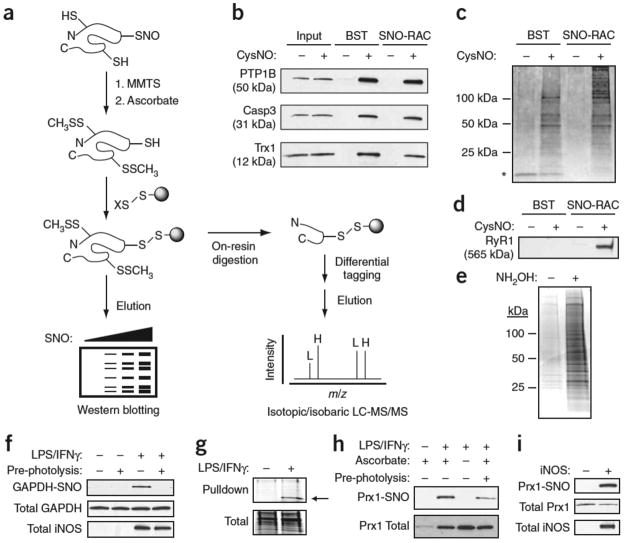
Figure 1. Analysis of protein S-nitrosylation by SNO-RAC.
(a) Protein thiols are blocked with S-methylmethanethiosulfonate (MMTS); ascorbate converts SNOs to free thiols; nascent thiols are covalently trapped with a resin-bound 2- or 4-pyridyl disulfide (denoted X); and proteins are eluted with reductant and analyzed by SDS-PAGE. Alternatively, proteins can be trypsinized on-resin to obtain peptides containing SNO sites, which may be differentially ‘tagged’ with isotopic or isobaric mass labels (L, light; H, heavy), and analyzed by MS. (b,c) HEK293 cells were incubated in the presence or absence of 0.5 mM CysNO for 10 min and subjected to a side-by-side comparison of the BST and SNO-RAC approaches (1 mg protein per sample). Proteins were separated by SDS-PAGE and visualized by western blotting (b) or silver staining (c) (see Supplementary Tables 1 and 6 online). Adventitiously eluted streptavidin is indicated by an asterisk. (d) Isolated rabbit sarcoplasmic reticulum (1 mg) was treated ±0.1 mM CysNO for 10 min, subjected to either the BST or SNO-RAC; western blotting was carried out to detect the 565 kDa ryanodine receptor 1 (RyR1). (e) Acyl-RAC was performed on HEK293-derived membranes (1 mg protein per sample). Proteins were visualized by SDS-PAGE and Coomassie staining. (f) RAW264.7 murine macrophages were treated ±0.5 μg/ml LPS and 100 U/ml IFNγ to induce inducible nitric oxide synthase, subjected to SNO-RAC (1 mg protein per sample) and analyzed as in c for S-nitrosylated GAPDH. Where indicated, extracts were photolyzed to verify assay specificity. (g–i) Macrophages were subjected to SNO-RAC (8 mg protein per sample) and eluants visualized by SDS-PAGE and Coomassie staining. The prominent band at ~22 kDa was excised and identified by MALDI-MS as peroxiredoxin-1 (indicated by arrow) (g). Lysates (1 mg protein per sample) from stimulated macrophages (h) and inducible nitric oxide synthase–transfected HEK293 cells (i) were subjected to SNO-RAC and analyzed for S-nitrosylated Prx1. For all experiments, n=3. Full-length scans are available in Supplementary Figure 7 online.