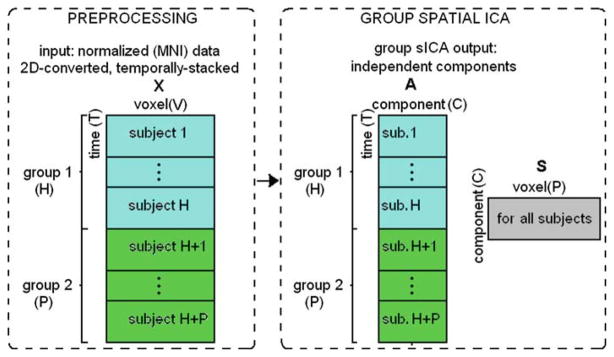
Fig. 4.
Summary of preprocessing and group spatial ICA steps. After each subject’s fMRI data are motion-corrected and spatially normalized to a common template, thus their voxels are aligned. The normalized data are flattened to a T-by-V matrix, where T is number of time-points and V is number of voxels. After normalization, H number of subjects for group 1 and P number of subjects for group 2 are temporally concatenated and the resulting (H + P)T -by-V matrix X is fed into group sICA algorithm. Group sICA finds C number of maximally independent (spatially) components. Each component’s spatial extent is described by C rows of the matrix S as a result of sICA. Each component has an associated time-course, for each subject (C columns of matrix A). X = AS