Figure 1.
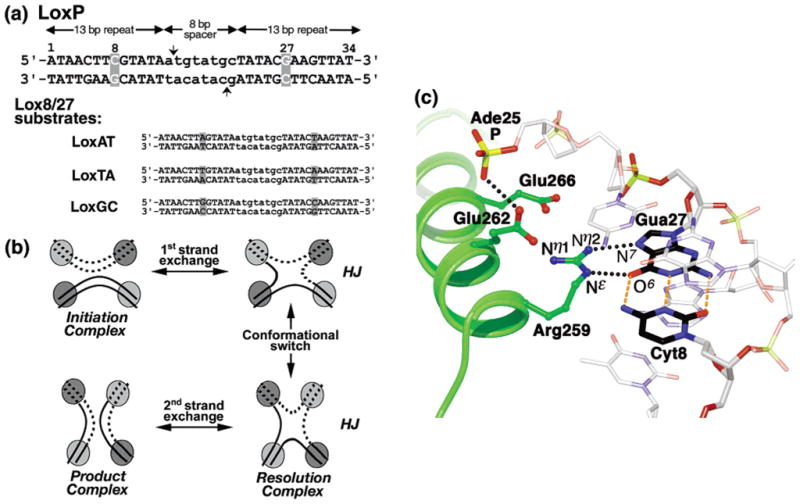
LoxP site, Cre-Lox recombination mechanism and the Arg259-Gua27 protein-DNA interface. (a) LoxP sequence and symmetric 8/27 substitutions. The 34 bp LoxP sequence is shown, with each position numbered 1–34 on the top strand. LoxP is comprised of two 13 bp inverted repeat binding elements (uppercase) surrounding an asymmetric 8 bp spacer (lowercase). Cleavage sites for strand exchange are indicated with vertical arrows. The position 8 and 27 substitution sites are highlighted (gray boxes). The 8/27 base pair substitutions are listed for the substrates LoxAT, LoxTA, and LoxGC. (b) Recombination mechanism. During recombination, four Cre monomers associate with two LoxP sites. Two Cre monomers are in an active “cleaving” conformation (light gray circles), while the other two monomers are in the inactive “noncleaving” conformation (dark gray circles). Strand exchange is accomplished by single strand cleavage to form a covalent phosphotyrosine-DNA intermediate (not shown) followed by strand “swapping” and religation of the homologous strands. The first strand exchange step (“initiation”) forms a Holliday junction (HJ) intermediate complex. During HJ complex isomerization, the Cre subunits undergo a “conformational switch”, in which inactive Cre monomers are now activated for cleavage, and the active monomers are inactivated. In the second strand exchange step (“HJ resolution”), the HJ is resolved to form recombinant products. We hypothesize that the Cre cleaving conformation has higher DNA affinity than the noncleaving conformation. (c) Protein-DNA interactions near the 8/27 substitutions, in a CreWT/LoxP HJ complex (35). Helix J (green ribbons) occupies the LoxP major groove, centered on the Cyt8-Gua27 base-pair (black bonds, interbase hydrogen bonds are indicated by orange dotted lines). Arg259 recognizes Gua27 O6 and N7 atoms via paired hydrogen bonds with the guanidinium Nε and Nη2 atoms, respectively, while Glu262 makes close contact (2.8 Å) with the phosphate of Ade25 (black dotted lines).
