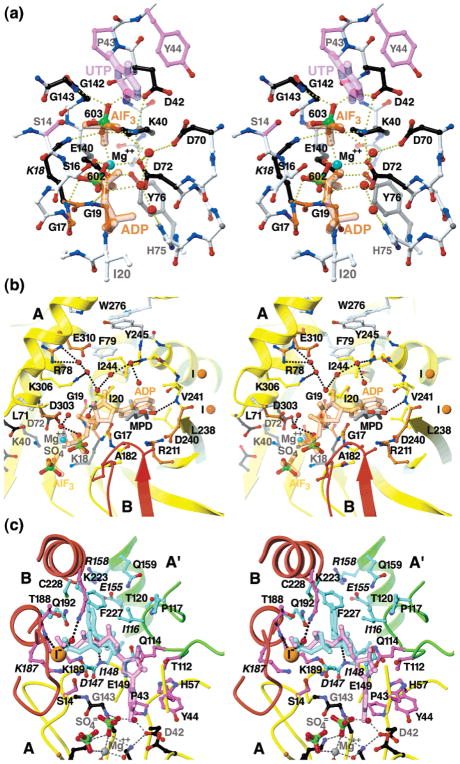
Figure 8.
Stereo diagrams showing the atomic details of the proposed (a) catalytic, (b) ATP, and (c) UTP/CTP subsites in the EcCTPS A subunit. The carbon atoms of conserved residues are colored-coded in ball-and-stick representations by their proposed functions: ATP binding (orange); UTP binding (purple); CTP binding (cyan); or catalysis (black). Hypothetical bound ligands, indicated by similarly color-coded translucent sticks, were placed as described in Experimental Procedures. Solvent molecules present in both A and B subunits are shown (red balls). Hydrogen bonds are indicated by the dotted lines. (a) The proposed catalytic subsite. For clarity, only the UTP uracil ring and C1′ atoms (purple), the ADP α- and β-phosphates, and AlF3 are shown (orange). Nonconserved residues (white) are shown are shown in ball-and-stick. Proposed catalytic residues Gly17, Lys18, Lys40, Asp70, Glu140, and Gly143 are conserved from DTBS. Asp72 occupies a similar position to Asn56 in DTBS. Asp42 is positioned to interact with the uracil O4 position. Hydrogen bonds for γ-phosphate binding and phospho/aminotransfer transition-state stabilization are potentially donated by the Lys18 and Lys40 amino and Gly143 amide nitrogen atoms. A putative magnesium ion (cyan ball), chelated by Asp72 and Glu140 and bridging the bound sulfate ions, directly overlaps the proposed activating magnesium ion in DTBS. The α- and β-phosphates would be recognized by the amides in P-loop residues 17–19, and bridging solvent interactions with Asp72 and His75. Conserved Tyr76 buttresses the active site through interactions with Glu140. (b) Proposed subsite for ATP. For the A (yellow) and B (red) subunits, backbone positions are indicated by ribbons and the nonconserved side chains are indicated by yellow ball-and-sticks. Additional conserved side chains are shown in ball-and-stick with white bonds. Positions of the magnesium ion (cyan), solvent molecules (red), and nearby iodine ions (“I”, orange) are indicated by balls. Hydrogen bonds are indicated by black dotted lines. ADP from the DTBS 1BS1 structure (translucent orange sticks) overlaps the EcCTPS-bound sulfate ions (green and red ball-and-sticks) and MPD molecule (gray and red sticks). By analogy with DTBS, the adenine recognition site is formed by the loop L8 (residues 240–245). (c) Proposed overlapping subsites for UTP substrate and CTP feedback inhibitor. The A (yellow) A′ (green), and B (red) subunit backbone positions are indicated by ribbons. Conserved side chains which line the proposed ligand binding surfaces are shown in color-coded ball-and-stick. For clarity, only the UTP subsite-proximal His57 conformation is shown. Positions of solvent molecules (red) and the iodine ion (“I–”, orange) are indicated by balls. Italicized labels indicate sites for which known change-of-function mutations exist (see text for details). The Lys187-Ala substitution abolishes ALase activity in EcCTPS. For the CTP site, some mutations, selected for resistance to CPEC or 5-fluorouracil, resulted in loss of CTP inhibition in CTPSs from C. trachomatis, Asp147-Glu, (56)), and hamster, Ile/Val116-Phe, Gly146-Glu, Ile148-Thr, Leu/Met151-Ile, Glu155-Lys, Arg158-His, and Asn/His229-Lys (57), where bold indicates a conserved residue for CTPSs, and the hamster residue is underlined. Residues Gly146, Ile151, and Asn229 are not shown for clarity. The sulfate/iodide site provide a potential site for binding the UTP or CTP γ-phosphates.