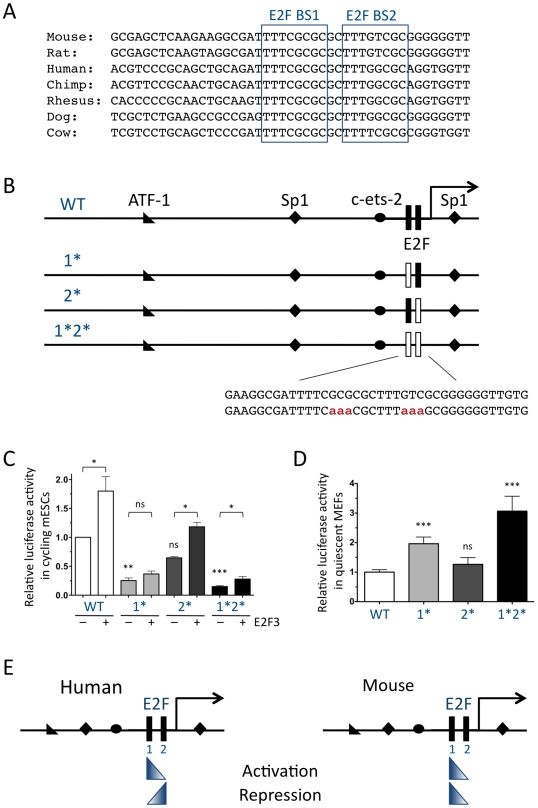
Figure 1. Regulation of the mouse p107 promoter through E2F binding sites in reporter assays.
(A) Conservation of the proximal p107 promoter across mammalian species. The two tandem consensus E2F binding sites (BS1 and BS2) are each indicated by a box. (B) Schematic representation of wild-type (WT), p107-1*, p107-2*, and p107-1*2* luciferase vectors. Transcription factor binding sites contained in this promoter region, as identified by sequence analysis, are indicated, as is the transcription start site (arrow). Black rectangular boxes indicate E2F consensus sites; white boxes indicate E2F consensus sites that are mutated. The inset represents the mutations (aaa) introduced in each site. (C) Relative luciferase activity expressed by the four constructs, co-transfected with CMV-E2F3 (+) or empty pCDNA (−), in cycling mESCs. For statistical analysis, each mutant construct was compared to the wild-type one and the effect of E2F3 on each construct was analyzed. (n = 3) (D) Relative luciferase activity in quiescent MEFs. (n = 15) (E) Comparison of the models for the regulation of the human and mouse p107 promoters by E2F based on reporter assays. Gradient triangles indicate the relative importance of each consensus E2F site to either activation or repression of p107.