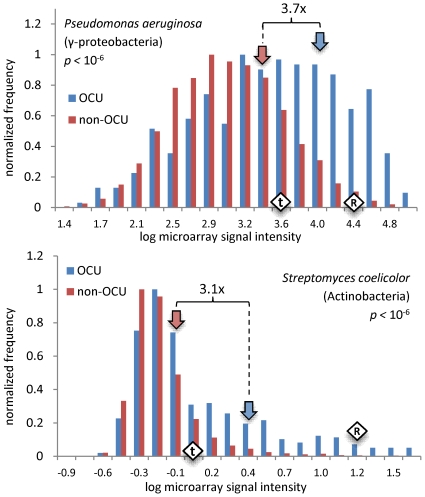
Figure 4. Expression levels of OCU versus non-OCU genes.
Histograms comparing microarray signal intensities between genes with optimized codon usage (OCU) and the non-OCU genes. The P. aeruginosa and S. coelicolor genomes were previously considered to lack translational selection (Text S1, Appendix B). The p-values are by the Baumgartner-Weiss-Schindler permutation test [43]. Block arrows show the mean microarray signal intensity of OCU or non-OCU genes. Numbers above the curly braces are ratios of mean signal intensity of OCU genes to mean signal intensity of non-OCU genes. Diamonds show the mean signal intensity for aminoacyl-tRNA synthetases (“t”) or the ribosomal protein genes (“R”). Full data for 19 organisms in Table S5; average ratio of OCU expression to non-OCU expression in the 19 organisms is 2.4x. See Figure S4 for similar histograms, but with the ribosomal protein genes excluded.