Figure 7.
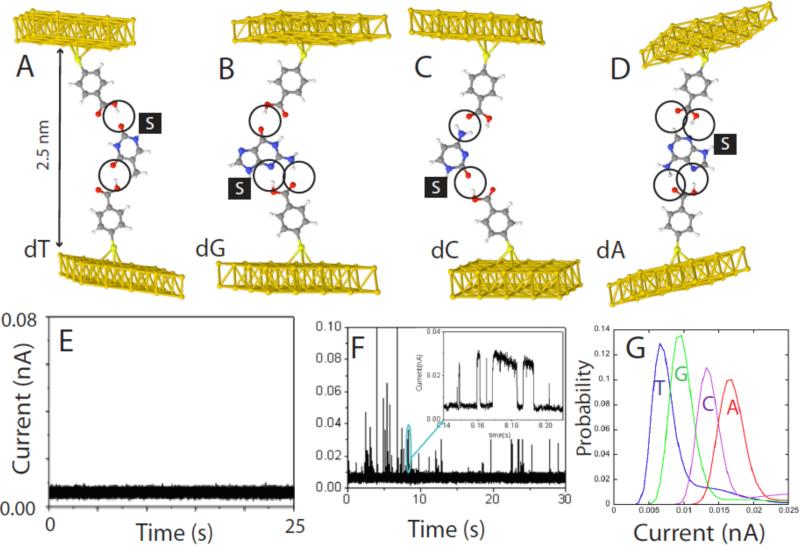
“Free-analyte” configuration of Recognition Tunneling for reading DNA bases. (A-D) show energy-minimized structures for the four nucleosides bound in a 2.5 nm gap with 4-mercapto benzoic acid as the reading reagent (R1, R2 in Figure 1C). The “S” stands for the deoxyribose sugar (not shown) and the order (dT, dG, dC, dA) corresponds to the predicted order of increasing tunnel conductance using density functional theory. (E) Shows the background tunnel current in organic solvent (trichlorobenzene) with the gap set to GBL = 12 pS (6pA at 0.5V bias). At this gap there is no indication of interactions between the two benzoic acid readers. (F) Shows an example of the current spikes that are observed when a solution of dG is injected into the tunnel junction. The inset shows details of some of the spike on a ms-timescale. Many of them show the telegraph noise switching characteristic of single molecule binding (the slight slope in the “on” level reflects the action of the servo used to control the tunnel gap). (G) Measured distributions of current for the four bases. The order agrees with the density-functional prediction, but the measured currents are larger than predicted. The overlap between reads limits the probability of a correct assignment on a single read to about 60%.