Abstract
Microbes are the most abundant and diverse organisms on Earth. In contrast to macroscopic organisms, their environmental preferences and ecological interdependencies remain difficult to assess, requiring laborious molecular surveys at diverse sampling sites. Here, we present a global meta-analysis of previously sampled microbial lineages in the environment. We grouped publicly available 16S ribosomal RNA sequences into operational taxonomic units at various levels of resolution and systematically searched these for co-occurrence across environments. Naturally occurring microbes, indeed, exhibited numerous, significant interlineage associations. These ranged from relatively specific groupings encompassing only a few lineages, to larger assemblages of microbes with shared habitat preferences. Many of the coexisting lineages were phylogenetically closely related, but a significant number of distant associations were observed as well. The increased availability of completely sequenced genomes allowed us, for the first time, to search for genomic correlates of such ecological associations. Genomes from coexisting microbes tended to be more similar than expected by chance, both with respect to pathway content and genome size, and outliers from these trends are discussed. We hypothesize that groupings of lineages are often ancient, and that they may have significantly impacted on genome evolution.
Symbiosis—as defined in its broadest sense (de Bary 1879; Saffo 1993)—is widespread in nature, ranging from obligatory mutualistic partnerships to commensalism to clearly detrimental, parasitic interactions (Paracer and Ahmadjian 2000). The phenomenon is not restricted to a particular domain of life, but can occur, for instance, between bacteria, archaea, and protists, which, in turn, can live together inside a specific animal host (Brauman et al. 1992; Tokura et al. 2000). Many instances of symbiosis are known, but they are not always understood mechanistically. The situation may not always be stable either: Symbionts may “cheat,” and/or compete among each other for a third partner (Palmer et al. 2003; Ferriere et al. 2007; Johnstone and Bshary 2008).
Leaving aside macroscopic organisms, symbiosis and local coexistence among single-celled microbes are even less well characterized. The extent, specificity, and stability of microbial associations are difficult to assess systematically in the environment, since elaborate staining procedures and/or molecular sequencing are needed in order to detect and differentiate between microbial lineages in situ. Nevertheless, several close partnerships between microbial species have already been identified. These include consortia of methane-oxidizing archaea and sulfate-reducing bacteria (AOM, “anaerobic oxidation of methane”) (Boetius et al. 2000; Caldwell et al. 2008; Knittel and Boetius 2009); consortia of phototrophic green sulfur bacteria surrounding motile beta-proteobacteria (Overmann and Schubert 2002; Wanner et al. 2008); consortia of sulfate reducers, sulfate oxidizers, and other lineages inside marine, gutless oligochaete worms (Dubilier et al. 2001; Woyke et al. 2006; Ruehland et al. 2008); and consortia of extremophilic lineages conducting ferrous iron oxidation in acidic pyrite mine run-offs (Tyson et al. 2004). Such groupings probably do not constitute “symbiosis” in a classical sense (Saffo 1993), but they are typically interpreted as syntrophic associations in which one partner consumes metabolites produced by the other. In addition, predatory and parasitic relationships are also known. An example for the latter is Nanoarchaeum equitans, a small archaeon that appears to be an obligate parasite of another archaeal species (Huber et al. 2002; Forterre et al. 2009). Despite such specific findings, the discovery of microbial associations has so far been largely interest-driven (or even fortuitous), meaning that a comprehensive picture of microbial coexistence has yet to emerge.
The notion that microbes in the environment perhaps exist in a less solitary manner than commonly assumed is also supported by the rapidly accumulating knowledge on intra- and interspecies microbial communication (Ryan and Dow 2008; Shank and Kolter 2009). Essential activities of single species such as nutrient uptake, biofilm formation, or cellular differentiation can be organized and synchronized by communication and cooperation (Parsek and Greenberg 2005; Waters and Bassler 2005; Kolter and Greenberg 2006; Gibbs et al. 2008; Ng and Bassler 2009). While it is less clear whether and to what extent microbes may interact with other species via specific communication, some bacteria are known to “eavesdrop” and to even respond to signals that they cannot themselves generate (Visick and Fuqua 2005). In addition, an interspecies relationship has been shown to evolve and quickly deepen in a laboratory evolution experiment (Hansen et al. 2007; Harcombe 2010).
Apart from the few cases of well-described, specific interactions, relatively little is known about how natural microbial assemblages form and how they are structured, if at all (Ruan et al. 2006; Horner-Devine et al. 2007; Fuhrman and Steele 2008; Raes and Bork 2008; Fuhrman 2009). They are often taxonomically highly complex and can encompass hundreds of different species, and at least some aspects of the composition of any given community are thought to be based on historical contingency (Martiny et al. 2006). Moreover, naturally occurring communities are difficult to reassemble and/or study under controlled conditions in the laboratory, since most of the constituting lineages are not available in pure cultures (typically more than 95% of species present in a given sample cannot be cultivated) (Staley and Konopka 1985). The difficulties in cultivating microbes have often been linked to slow growth and unknown nutritional requirements, but might also be partly attributed to their synecology—for example, reflecting a need to coexist within a biofilm or to aggregate together with partner species in order to ameliorate adhesion (Min and Rickard 2009) or to dispose of otherwise inhibitory metabolic products.
Since the establishment of the first comprehensive microbial phylogeny using the 16S rRNA gene (Fox et al. 1980) and the invention of techniques for rapidly generating large blocks of 16S rRNA sequence data (Lane et al. 1985; Giovannoni et al. 1990; Ward et al. 1990), a great variety of environments have been sampled to study microbial diversity in situ. Today, the 16S rRNA gene remains the marker of choice for identifying microbes in their environments, and the size of databases dedicated to this gene is growing exponentially (Desantis et al. 2006; Pruesse et al. 2007; Cole et al. 2009). In addition, environmental sequences are increasingly being annotated with contextual information (e.g., geographic position, temperature). An important effort to define and standardize such sequence meta-data has been initiated within the Genomic Standards Consortium (Field et al. 2008a,b), specifically by developing the MIENS standard (minimum information about an environmental sequence). However, the existing annotations of many of the legacy sequences in the databases will have to be migrated to such standards, which requires considerable effort. The results of such efforts are increasingly being made available in integrated resources such as CAMERA (Seshadri et al. 2007), IMG/M (Markowitz et al. 2008), and megDB (Kottmann et al. 2010), but at present only a small minority of 16S rRNA sequences have geo-referencing or other contextual information.
Using 16S rRNA sequences in combination with other data, classical ecological questions including species (co)-occurrence and diversity have also been addressed extensively in microbes (Bell et al. 2005; Langenheder et al. 2006; Ruan et al. 2006; Horner-Devine et al. 2007; Smith 2007; Langenheder and Prosser 2008). In doing so, many of the concepts that have originally been developed for macroscopic organisms have been adapted and applied to microbes. However, these studies have mostly focused on one specific environment, or one specific lineage, at a time (e.g., Alonso et al. 2007; Newton et al. 2007; Fuhrman and Steele 2008) (this way, ecological questions can be studied in a more defined setup). What has not been addressed much, so far, is the global partitioning of microbial lineages among all sampled environments. Here, we take a first step in this direction, by systematically studying a current snapshot of the complete data set of full-length 16S rRNA sequences. We search for groups of lineages that occur together more often than expected by chance, and we connect this information to genomic data, as well as to the limited metadata that are available regarding the sampling sites (the latter information stems mostly from free-text annotations provided at the time of database submission). We find that the assortment of lineages and environments is clearly nonrandom, and that specific and recurring associations among lineages can be described, at various levels of detail and phylogenetic resolution.
Results and Discussion
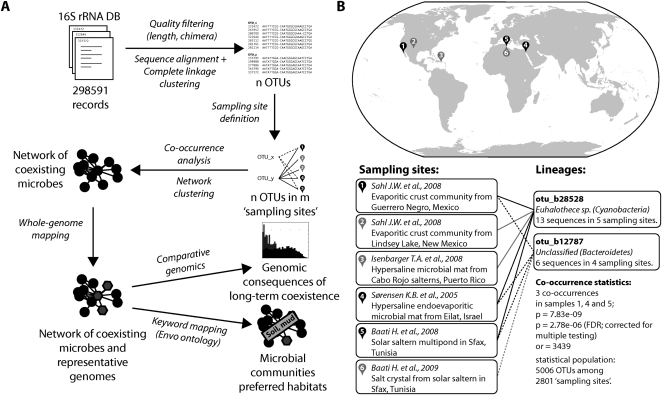
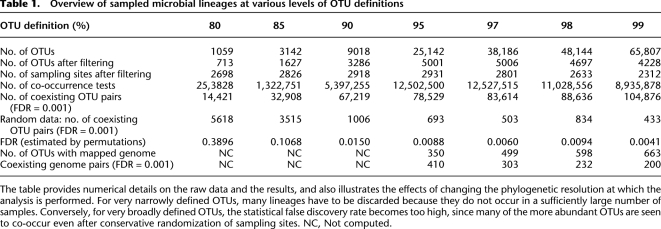
In order to comprehensively characterize the occurrence of microbial lineages in the environment, we first grouped publicly available, full-length 16S rRNA sequences at various levels of sequence identity, thereby creating unsupervised sets of “operational taxonomic units” (OTUs; see Methods for details). Each OTU was assigned a taxonomic annotation that reflected the consensus of its member sequences, and a single sequence was chosen to represent each OTU in subsequent sequence comparisons. Next, we comprehensively compiled environmental “sampling events” of 16S sequences; such an event is defined here as a unique combination of submitting authors, project title, and isolation source, as annotated in the respective database records. We assumed that sequence entries for which all three fields are exactly identical were sampled together, at a given site. Our procedure (Fig. 1) thus resulted in a large matrix that connects OTUs to environmental sampling events (Table 1). Depending on the OTU definition, this matrix contained roughly between 700 and 5000 distinct OTUs, which were mapped to roughly 3000 distinct sampling events (we only retained sampling events that encompassed at least two OTUs, and conversely, only OTUs that were observed in at least three sampling events).
Figure 1.
Detection of coexisting microbial lineages. (A) Schematic description of the analysis procedure. Publicly available 16S ribosomal RNA sequences are first grouped into operational taxonomic units (OTUs), then annotated according to unique environmental sampling events, and finally searched for statistically significant co-occurrences. Where available, completely sequenced genomes are mapped onto the resulting network, which is then clustered and annotated. (B) Example for a specific lineage association. The two lineages (defined at a 97% 16S sequence identity cutoff) have been sampled overall relatively rarely, but they occurred together three times, at three distinct sites. (or) Odds ratio. Under “Sampling sites,” the investigative work of “Baati H. et al., 2009” refers to R Amdouni, E Ammar, H Baati, N Gharsallah, and A Sghir (unpubl.).
Table 1.
Overview of sampled microbial lineages at various levels of OTU definitions
The table provides numerical details on the raw data and the results, and also illustrates the effects of changing the phylogenetic resolution at which the analysis is performed. For very narrowly defined OTUs, many lineages have to be discarded because they do not occur in a sufficiently large number of samples. Conversely, for very broadly defined OTUs, the statistical false discovery rate becomes too high, since many of the more abundant OTUs are seen to co-occur even after conservative randomization of sampling sites. NC, Not computed.
Next, we examined this matrix for any non-random assortment of OTUs to environments, which would manifest itself as groups of OTUs observed together more often than expected by chance. Our underlying null model is that of global, random dispersal of lineages across environments (Harvey et al. 1983; Finlay 2002; Kunin et al. 2008a; Hubert et al. 2009), and essentially corresponds to the first part of Baas Becking's enigmatic statement, “Everything is everywhere, but, the environment selects” (de Wit and Bouvier 2006). While this null model is clearly not applicable for macroscopic organisms with distinct biogeographic distribution patterns, it does represent the simplest default assumption for microbes, and it is appropriate for the very large geographical and temporal scales that we consider here. By computing the hypergeometric probability of pairwise co-occurrences and correcting for multiple testing, we found that, indeed, a large number of statistically significant associations between OTUs can be observed, irrespective of the precise choice of OTU definition cutoff (Fig. 2; Supplemental Fig. S1; Table 1). A concrete example for such an association is shown in Figure 1B (data from Sorensen et al. 2005; Baati et al. 2008; Isenbarger et al. 2008; Sahl et al. 2008; R Amdouni, E Ammar, H Baati, N Gharsallah, and A Sghir, unpubl.): a well-characterized lineage of Cyanobacteria (belonging to the halophilic Euhalothece) (Garcia-Pichel et al. 1998) was observed to be associated with an uncharacterized lineage having no cultivated or named representatives (a monophyletic sister group of the Psychroflexus lineage [Bacteroidetes]). This particular association is based on three independent sampling events in which both lineages had been observed together, by three distinct laboratories in three distinct countries. Considering that the OTU definition in this case is relatively narrow (97%) and that this association occurs against a backdrop of about 2800 sampling events covering more than 5000 OTUs, the observation becomes highly significant (P < 3 × 10−06; after multiple testing adjustment). Overall, several thousand of such associations could be identified. To assess the effects of potential biases in the sampling data, and in order to estimate our false discovery rate (FDR) empirically, we performed a conservative randomization of our data—by keeping constant the size distributions of both sampling events and OTUs, but shuffling the connections between OTUs and sampling sites. This resulted in a reduction of the number of reported associations by >99% for most of the OTU definition cutoffs (Table 1), which translates to FDRs of ∼1%, except at very broad OTU definitions (i.e., when setting the OTU clustering cutoff to 85% sequence identity or less) (see Table 1). The few remaining false-positive associations were observed mainly among widely sampled lineages known to occur inside the mammalian digestive tract; this likely reflects the strong study bias toward 16S gene sequences of this habitat (Ley et al. 2008; Hamady and Knight 2009).
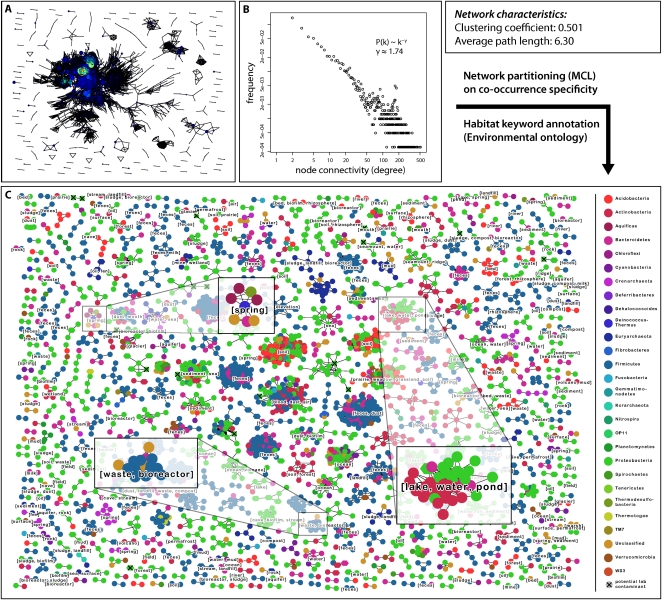
Figure 2.
Global network of coexisting microbial lineages. (A) Overview of the network of lineage associations. Each node denotes a microbial lineage, and each line a significant co-occurrence relationship. Node size is proportional to the number of sequences in the lineage, and node color indicates the connectivity degree of a node (along a color gradient: blue, low connectivity; red, high connectivity). Throughout the figure, the OTU definition cutoff is at 97% sequence identity, and the P-value cutoff for an association is 0.001 (i.e., FDR after correction for multiple testing). (B) Connectivity degree distribution plot for the network in A. The distribution is coarsely compatible with a power law distribution. (C) Same network as in A, but partitioned using unsupervised Markov clustering, to reveal modules (clusters) of co-occurring lineages. Here, node color denotes taxonomic classification at the phylum level. Lineages suspected to contain potential laboratory contaminants (Tanner et al. 1998; Barton et al. 2006) are mainly observed in small clusters, and are marked with a small black X (17 such lineages in total).
In addition to assessing the statistical significance, we also computed a “specificity” value (or “association strength”) for all OTU pairs. This value corresponds to the Jaccard similarity; it is 1.0 if a pair of OTUs is always observed together (but never separately), and zero for a pair of OTUs that is always observed in distinct environments, but never together. Remarkably, we found associations at both extremes of specificity (i.e., close to 1.0 or close to zero) (see Supplemental Fig. S3 for the overall distribution). An example for the former is shown in Supplemental Figure S1A: a previously undescribed bacterial OTU (a sublineage of the candidate division JS1) was observed together with a specific Methanosarcinales lineage in marine sediments, again by three distinct laboratories (once in the Mediterranean, twice in the Gulf of Mexico at distinct sites). However, in this case, the two lineages were never found separately, in any of the 2800 environment samplings we studied. The likelihood of observing such a specific association by chance is again very low (P = 1.1 × 10−7 after correction for multiple testing). An example for a less specific but nevertheless highly significant association is shown in Supplemental Figure S1B: A lineage of gamma-Proteobacteria (nosocomial pathogens from the genus Stenotrophomonas) was frequently observed together with a lineage of Bacilli (genus Staphylococcus). The two lineages were sampled together 10 times—by seven distinct laboratories—in various air samples, skin samples, dust, and on Chinese cabbage. The association is highly significant (P < 10−9 after correction for multiple testing), but less specific: Both lineages have also been observed separately (in 32 and 17 sampling sites, respectively). Not all of the latter observations were related to skin samples. Stenotrophomonas, for example, may also form distinct blooms in shallow coastal lagoons (Piccini et al. 2006). While co-occurrence alone cannot offer any mechanistic explanation for lineage associations, the additional information in the specificity of an association does provide a constraint when discussing possible scenarios (obligatory mutualism, for example, would be expected to result in a high association specificity). Barring any additional information, we did choose to interpret our observed associations conservatively, by assuming that they for the most part simply reflect shared or overlapping niche preferences. Instances of undescribed, specific mutualisms and parasitisms are presumably contained within our findings, but additional experimental follow-ups will be required for a detailed characterization of such interactions (Orphan 2009). That notwithstanding, this first part of our analysis already provides an empirical base for discovery and allows us to explore more specific hypotheses about the reasons for the coexistence of sets of uncultured genotypes.
Next, we searched our observed co-occurrence relations for previously known microbial associations (Supplemental Fig. S1). While we did not recover the known association between the Nanoarchaeum and Ignecoccus lineages, nor the Chlorochromaticum consortium, we did find strong evidence for AOM consortia (Supplemental Fig. S1D). We also observed the known association between the lineages Leptospirillum (phylum Nitrospira) and Acidothibacillus (phylum Proteobacteria), both of which are known to thrive in acidic bioleaching environments. In this case, the association we found was remarkably strong and specific: Out of 21 independent observations of Leptospirillum (by 18 distinct author teams in various settings), all but a single one also included observations of Acidothiobacillus (i.e., 20 out of 21; P < 10−35) (Supplemental Fig. S1C) (in this case, the OTU clustering distance was 90%). Remarkably, this association appeared to be somewhat asymmetric: Acidothiobacillus did occur occasionally without its partner (in an additional 18 sampling events), suggesting that the mutual dependencies might not be equally strong in both directions. As a further test of our associations, we conducted an independent co-occurrence search of microbial lineages in the published literature (Supplemental Fig. S5). The frequencies of co-mentions of species names in PubMed can, indeed, reveal ecological associations (Freilich et al. 2010), albeit limited to those lineages that are already validly named and for which cultivated type strains typically exist. We find that more than 70 of our pairwise associations (counting nonredundantly at the genus level) can, indeed, be confirmed by the published literature, that is, their co-mention counts rise above a conservative randomization of species names and PubMed entries (Supplemental Fig. S5). Apart from the known associations, we also observed a large number of previously undescribed interactions, many of which involved unclassified lineages without any cultured or named representative (discussed below; the full set of associations is also available for browsing online). It should be noted that our data set likely misses some aspects of microbial coexistences, due to experimental biases in the generation of 16S rRNA sequences. In particular, the frequent choice of primers that will not target archaeal sequence types (Muyzer et al. 1995) in environmental studies may lead to an underestimation of the association between bacteria and archaea (but see Supplemental Fig. S1 and Fig. 5, below, for examples).
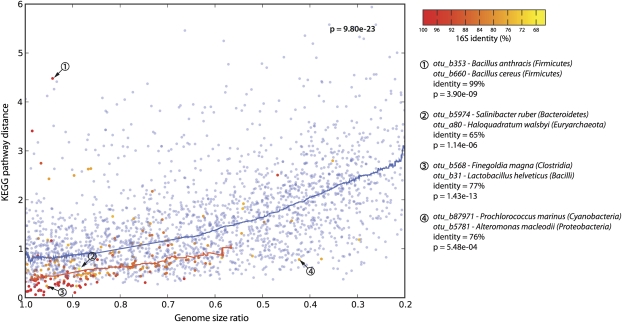
Figure 5.
Functional similarities among co-occurring genomes. Each dot denotes a pair of genomes, which are either co-occurring in the environment (red to orange dots) or randomly paired (blue dots). The plot shows differences in functional genome content (y-axis), and in genome size (x-axis). Lines denote running medians. Note that, in general, the more divergent two genomes are in size, the more they are functionally distinct (blue line). In co-occurring genomes, this trend is strongly shifted toward similar functions, at all levels of phylogenetic relatedness (color-coded from red to orange). Examples of genome pairs that are discussed in the text are indicated.
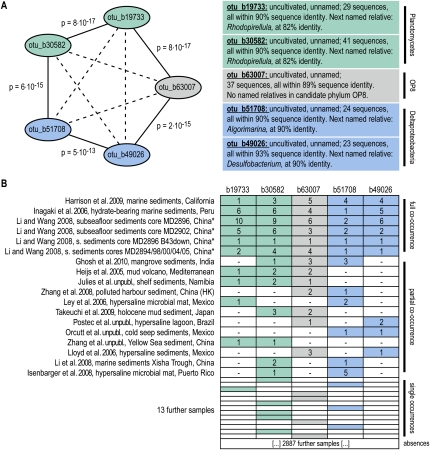
The observed associations were not limited to pairwise co-occurrences. When plotting the associations as a graph, a densely connected network of OTUs emerged (Fig. 2A). The topology of that network is clearly nonrandom; it exhibits a high clustering coefficient, short average minimum path length, and a connectivity degree distribution that has no characteristic maximum (i.e., the network is roughly matching the “scale free, small-world” criteria) (Barabasi and Oltvai 2004). This topology suggests that the network can be meaningfully partitioned, and that doing so should reveal modules of densely connected microbial lineages; these might be regarded as the microbial equivalents of the “syntaxa” of vegetation analysis. One such possible partitioning is shown in Figure 2C; it conveys more information than a simple list of pairwise co-occurrences because it groups specific lineages, at the exclusion of others. Module formation can occur even if the various pairwise correlations are not all highly significant (e.g., due to undersampling); this is because a certain fraction of missing or poorly scoring associations can be tolerated as long as the overall topology remains that of a tightly linked module. Furthermore, partitioning allows the annotation of keywords that describe the commonalities among the associated sampling sites of the various modules (Fig. 2C; see Methods). Among the modules, we observed intriguing cases where all or the majority of lineages have not been characterized before. An example is shown in Figure 3 (data from Heijs et al. 2005; Inagaki et al. 2006; Ley et al. 2006; Lloyd et al. 2006; Isenbarger et al. 2008; Li and Wang 2008; Li et al. 2008; Zhang et al. 2008; Harrison et al. 2009; Takeuchi et al. 2009; Ghosh et al. 2010)—five lineages that are co-occurring very specifically in certain marine sediments; they are from three distinct phyla, and each lineage is entirely uncharacterized. (A closer phylogenetic analysis revealed that the two Planctomycetes OTUs are related to each other, to the exclusion of other Planctomycetes lineages; they have been found also in other marine and freshwater environments, and our co-occurrence thus defines a more restricted home context for this lineage.) Specific modules such as this example are striking and likely provide a first glimpse onto hitherto undescribed microbial consortia.
Figure 3.
Example of a novel, previously undescribed module of coexisting lineages. (A) Five distinct microbial lineages are shown; they belong to three different phyla and are defined at an OTU-clustering distance of 90% sequence identity at the 16S rRNA gene. The five lineages have been exclusively observed through environmentally sampled sequences and have not been named. (B) The table shows all occurrence counts of these lineages among our sampling data; the P-values indicated have been corrected for multiple testing, against the background of all lineages defined at 90%. Adjusted P-values (FDR; p) and odds ratios (or) are indicated. (*) The samples by Li et al.(2008) have been collected at distinct sites, covering a distance of more than 600 miles; collection was at different water depths and sampling dates. Investigators involved in unpublished work are as follows: E Julies, V Bruechert, and BM Fuchs; B Orcutt, SB Joye, S Kleindienst, K Knittel, A Ramette, A Rietz, V Samarkin, T Treude, and A Boetius; A Postec, R Warthmann, C Vaconcelos, K Hanselmann, and J McKenzie; Z Zhang, H Xiao, and X Tang.
While our 16S-based OTUs provide fairly objective coverage of phylogenetic lineage space, they do not, in themselves, contain any information about molecular and ecological functions. We therefore attempted to represent each OTU by its best match among completely sequenced genomes, to the extent that the latter are available (see Methods). Strains for which complete genomes have been sequenced do not usually originate from the environmental samplings described here. However, as long as they are closely related to the OTU in question, they may suffice to reveal broad genomic trends related to coexistence. The validity of this approach is based on two observations/assumptions. First, our co-occurrence analysis is already enriching for lineages that are likely abundant (Pedros-Alio 2006) and that can be widely found and easily accessed (each OTU had to be sampled at least three times to be included here). And, second, there seems to be a notable stability of environmental habitat preferences among microbial lineages in general (Von Mering et al. 2007; see also below). This suggests that a sequenced strain may represent other members of its OTU in terms of its genomic content and aspects of its ecology even if it has diverged from them to some degree. We were able to map between 350 and 660 genomes to a subset of our OTUs (this depends on the OTU resolution; notably it also means that a significant fraction of sequenced genomes currently cannot be connected to an OTU that has been repeatedly observed in the environment). This mapping translates to between 200 and 410 significant partnerships for which genomic information is available for both partners, covering a small but significant fraction of all the instances of co-occurrence we detected. To our knowledge, this is the first time that a global, environmentally motivated association network between genomes has been constructed.
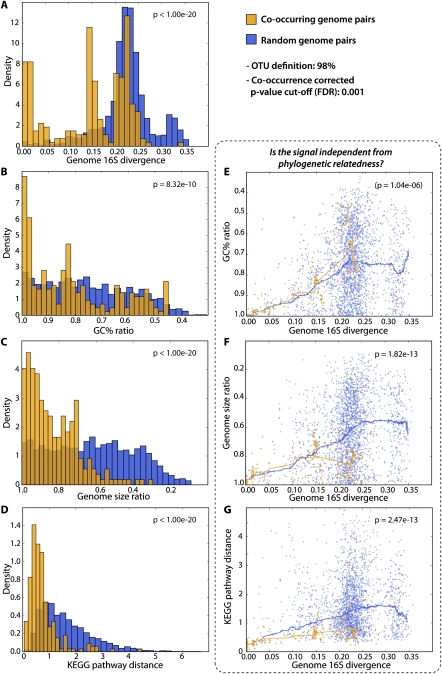
We used this network to objectively assess potential constraints on genome evolution, which might be a consequence of the association of a genome to its preferred environment and to other lineages in that environment. We observed four highly significant trends among co-occurring genomes: They tend (1) to have more similar genome sizes, (2) to be more similar in GC content (i.e., the fraction of the genome consisting of guanine and cytosine), (3) to be more similar with respect to relative coverage of functional pathways, and (4) to be phylogenetically more closely related than randomly selected pairs of genomes (Fig. 4). The latter trend was also visible from 16S sequences alone (Supplemental Fig. S2). The trend to phylogenetic relatedness is presumably the easiest to rationalize: Pairs of ecologically associated lineages, which are also closely related phylogenetically, would arise naturally assuming that neither lineage had changed their habitat preferences since they split from their last common ancestor. We indeed observe this signal and detect that it extends surprisingly far back in time: Lineages that have diverged up to 10% at the 16S sequence identity level are still clearly enriched among environmentally associated pairs (Fig. 4A; the peak seen at 15% sequence divergence is largely due to a single, well-covered cluster; see Supplemental Figs. S6, S8). In principle, this relatedness signal could also explain our three other observations: Phylogenetically related genomes are known to exhibit similar GC contents, genome sizes, and functional composition. To assess this possibility, we tested these three signals for independence from the phylogenetic signal, by correcting for the underlying correlations as learned from randomly selected genome pairs (Fig. 4). In the case of GC content similarity, we find that the signal can, indeed, be largely explained by phylogenetic relatedness alone—it is not an independent observation. This would argue against environmental selection on GC content, at least at longer time scales, and it gives further support to algorithms that partition environmental sequences based on genomic signatures (McHardy and Rigoutsos 2007; Mrazek 2009). In contrast, importantly, we observed that both genome size similarity and functional similarity could not be explained solely by phylogenetic relatedness. For example, while randomly selected pairs of genomes have genome sizes that can vary considerably, environmentally associated genome pairs tend to level off at ∼20%–30% genome size difference, on average (Fig. 4F, P < 10−13). This is remarkable because it suggests that a given environment tends to select for a particular optimal genome size range, even across distinct lineages; furthermore, it suggests that lineages spend sufficient time in their preferred environments to allow for these optimal genome sizes to be selected for and maintained (against a mutational spectrum that is thought to be largely biased toward deletions in bacteria) (Mira et al. 2001; Nilsson et al. 2005). Our observation confirms what has been known anecdotally from a number of environments: Planktonic marine environments, for example, persistently select for small to very small genome sizes (Giovannoni et al. 2005; Ting et al. 2009), whereas soil microbes are often among those with the largest genomes. Our results are also in line with observations indicating different average genome sizes in distinct environments (Raes et al. 2007; Angly et al. 2009). Regarding the functional similarity of genomes, we likewise observe that it is much stronger than what would be expected based on relatedness alone (Fig. 4G). Here again, lineage-environment associations appear to be stable enough to allow selection for similar functional repertoires even in unrelated lineages.
Figure 4.
Coexisting lineages display similarities in genomic features. Here, we exclusively focus on co-occurring lineages for which completely sequenced genomes could be mapped to both partners (this genome mapping is globally visualized in Supplemental Fig. S6). Properties of such co-occurring genomes are compared, and contrasted against randomly paired genomes. (A) The distribution of 16S sequence divergence scores; shifted to the left for co-occurring genome pairs (i.e. they tend to be related phylogenetically). In panels E, F, and G, we test for independence between phylogenetic relatedness, and observations as shown in panels B, C, and D, respectively. Here, each dot denotes a pairwise genome comparison, and lines correspond to running medians.
However, apart from a phylogenetic signal, functional similarities can also arise due to similarities in genome size (van Nimwegen 2003; Konstantinidis and Tiedje 2004; Ranea et al. 2004). When correcting for the dependency between genome size and functional content, we again find that co-occurring genomes of identical size are much more similar in functional terms than expected (Fig. 5). In Figure 5, we not only plotted genome size and functional similarity, but also phylogenetic relatedness (by means of a color code). This reveals the expected, combined trends: Environmentally associated lineages that tend to be most similar in functional terms also tend to be those that are both, phylogenetically the most related and also the most similar in terms of genome size. Outliers from these trends should reveal interesting exceptions, inviting speculations on distinct ecological scenarios. We highlight a few of such extremes in Figure 5. The first example represents an outlier case because the two lineages are very closely related phylogenetically (both are Bacilli), yet they currently hold the record in terms of functional divergence. Note that the various species of Bacilli are very closely related phylogenetically and cannot easily be distinguished based on 16S alone (Vilas-Boas et al. 2007; Kolsto et al. 2009) (our algorithm thus assigned the two genomes arbitrarily among the two lineages, well within 98% sequence identity). Nevertheless, this reflects on the known, large phenotypic and genomic diversification within the so-called Bacillus cereus “group” (Vilas-Boas et al. 2007; Kolsto et al. 2009) (e.g., Bacillus anthracis is usually nonmotile and produces a capsule and toxins, whereas B. cereus tends to be motile and to make no capsule). Importantly, our data show that such a high level of phenotypic and genomic plasticity among co-occurring lineages is exceptional, especially when they are so closely related phylogenetically. Perhaps the unusual life cycle of Bacilli (involving a resilient endospore stage) is conducive to unusually large changes in lifestyle and phenotypes, over short time periods. In contrast, the second example describes two lineages that are very distant phylogenetically (one is an Archaeon and the other a Bacterium), and yet they co-occur quite specifically. In our data, these two (Salinibacter and Haloquadratum) are outliers because, despite their distance, they have very similar genome sizes and very similar functional pathway coverage, marking the current record at such a large phylogenetic distance. Perhaps, both lineages have independently entered the same niche (i.e., warm, fully oxygenated brines) (see also Kunin et al. 2008b), and have thus converged toward coarsely similar overall genomic features. The next example again presents two fairly unrelated lineages (related only at the phylum level), which are even more closely matched in terms of genome size and functional genome composition. They occur together very specifically, on acidic human skin, dust, and in filtered air (seven observations by five distinct laboratories; P = 6 × 10−13; note that dust and air samples are related to skin since they may contain small skin-derived particles). Despite apparently occupying the same niche, it is notable that only one of them (Finegoldia) has a tendency for opportunistic pathogenesis (Goto et al. 2008). The last example concerns two lineages that occur together in the open ocean (Prochlorococcus and Alteromonas). They are unrelated phylogenetically, and they were chosen as outliers here because their genomes are unusually distinct in size (Alteromonas is more than twice as large as Prochlorococcus) (Rocap et al. 2003) (note that our analysis has insufficient resolution to specify the exact “ecotype” for either lineage). Since both were sampled in the open water, it is difficult to envisage any mechanism of their association, but this particular pair has been noted before—Alteromonas has been enriched as a co-contaminant in Prochlorococcus cultures, facilitating the growth of the latter (Morris et al. 2008) (perhaps by alleviating oxidative stress). In the ocean, the association is probably rather unspecific, although it is conceivable that Alteromonas, as an opportunistically growing heterotroph, may profit from biomass accumulated by the primary producer Prochlorococcus. These lifestyles are quite distinct and might explain the unusually large differences in genome size.
Overall, however, we find that co-occurring genomes tend to closely match each other's genome sizes and broad functional composition. These results seem to be compatible with a picture of competition (Hibbing et al. 2010), rather than cooperation, among most of the distinct microbial lineages found at any given site: If the majority of lineages were to routinely cooperate by specialization and division of tasks, this would presumably result in genomic features that might become more distinct from each other over time. Of course, the broad view that we take here could easily make us miss cooperation among a subset of lineages, such as syntrophy and other mutual benefits from the juxtaposition of distinct molecular capabilities. But such interactions are perhaps anyway more fleeting encounters rather than stable mutualisms. Indeed, long-term obligatory mutualism usually requires stable and specific physical contact between the organisms in question (Boucher 1985), a requirement that makes it perhaps less feasible for microbes that are generally dispersed easily (except, of course, when vertically inherited together within a eukaryote) (Vautrin and Vavre 2009).
In the future, statistical approaches like ours stand to benefit greatly from the projected further increases in both microbial genome sequencing (Ahmed 2009; Chain et al. 2009; The NIH HMP Working Group 2009) and 16S rRNA sampling (Tringe and Hugenholtz 2008; Costello et al. 2009). Both types of data will prove particularly valuable when augmented with standardized information about the environments sampled, for example, by following the recommendations of the MIENS standard (http://gensc.org/gc_wiki/index.php/MIENS). Novel and specific microbial assemblages can already be identified using the current data (see Fig. 3; Supplemental material), and more such discoveries can be expected with higher data coverage. Note that our approach does not require prior information about environmental ontologies or hierarchies of sampling sites; instead, groups of biologically related sampling sites are defined by the data themselves (Fig. 2; Supplemental Fig. S11). In general, approaches that integrate sequence data from both strain sequencing and from environmental marker gene sequencing hold great potential, since they connect the molecular information contained in the (pan-)genome of each lineage to the quantitative occurrence pattern of that lineage around the globe.
Methods
Definition of taxonomic units and sampling events
All 298,591 available 16S rRNA sequence records were downloaded from the Greengenes database (Desantis et al. 2006) on January 2009. At Greengenes, these sequences had already been cleaned of potential chimera by the program Bellerophon (Huber et al. 2004). We filtered sequences according to their lengths (≥900 nt for Archaea and ≥1200 for Bacteria) and additionally flagged sequences predicted to be chimeric by the program ChimeraSlayer (http://microbiomeutil.sourceforge.net/). We also removed from the analysis all sequences lacking annotations in any of the fields “author,” “title,” or “isolation_source.” This was done in order to be able to define a sampling event for each record. In our study, a “sampling event” is defined as the unique concatenation of these three annotation fields (author + title + isolation_source).
Archaeal and bacterial sequences were aligned separately, using the secondary-structure aware aligner “Infernal” (Nawrocki et al. 2009), together with the corresponding 16S rRNA covariance models of the RDP database (Cole et al. 2009). Before defining OTUs, we removed sequences for which the alignment had not been successful (i.e., Infernal bit-score < 0). OTUs were built for both Archaea and Bacteria by hierarchical clustering (complete linkage), at various distances (from 0.2 to 0.01), using the clustering tool of the RDP pyrosequencing pipeline (Cole et al. 2009; http://pyro.cme.msu.edu/). Because not all 16S sequences reported in databases are necessarily genuine environmental sequences (Tanner et al. 1998; Barton et al. 2006), we assembled a database of potential laboratory contaminants, containing 38 distinct sequences (Tanner et al. 1998; Barton et al. 2006). Homology searches revealed that between 47 and 309 of our OTUs contained such sequences (matching at 97% sequence identity or better). However, these OTUs are rarely involved in significant co-occurrences; for example, in Figure 2 only 17 of the OTUs shown contain potential contaminants, and these are scattered over various smaller clusters (they are flagged in Fig. 2 and in the detailed Supplemental material).
In order to compute sequence divergence values for pairs of OTUs, we first selected a single sequence to represent each OTU. (We chose the sequence that had the minimum sum of squares of distances to all other sequences within that cluster; note that this does not favor short sequences since the distances we used are length-normalized.) We then aligned these representative sequences pairwise (using the program “water” from the EMBOSS package) (Rice et al. 2000) and determined their sequence identity.
Classification of taxonomic units
In order to assign taxonomic classifications to entire OTUs, we first assessed the pre-annotated taxonomies of all individual 16S rRNA sequences in Greengenes (sensu RDP taxonomy). Where these were still annotated as “unclassified,” we re-ran the taxonomy classification using the RDP classifier (Cole et al. 2009). Taxonomy predictions reported there were considered reliable, if supported by a minimum bootstrap value of 80%. To assign taxonomy classifications to OTUs, we then used a simple majority vote: If more than half of the sequences present within a cluster agreed upon a classification, the OTU was annotated as belonging to this taxon. In case of conflicts, we assigned consensus classifications at increasingly higher levels of taxonomy until the majority vote condition was again met.
Co-occurrence analysis
In order to reduce the search space for co-occurrence testing (which encompasses potentially more than 2 billion pairs, for example, in the case of OTUs defined at 99% sequence identity), we limited our analysis to OTUs occurring in at least three distinct sampling sites. Conversely, we only considered sampling events encompassing at least two distinct OTUs. For these “filtered” OTUs and samplings (see also Table 1), we tested the co-occurrence significance for all possible pairs using the Fisher's exact test. For each test, the four cells in the contingency table denoted the number of samples containing both OTUs, one of the two OTUs only, or none of the two, respectively. Subsequently, we adjusted all P-values for multiple testing using the Benjamini and Hochberg FDR controlling procedure (Benjamini and Hochberg 1995), as implemented in the “multtest” library of the statistical software package R (http://www.r-project.org). We also verified our FDR empirically, by re-computing the associations using randomized input data. For this, we randomly reassigned the various OTUs to the various sampling events, under the constraint that each OTU kept the overall number of samples it mapped to, and each sample kept the overall number of OTUs. This maintained the size distributions of both, samples and OTUs (results are provided in Table 1). To compute the necessary large number of tests in a reasonable time, we used a C-implementation of the test in the Apophenia library for scientific computing (http://apophenia.sourceforge.net), using the python SWIG interface as a wrapper. For selected examples of co-occurring lineages discussed in the text (Figs. 1, 3; Supplemental Fig. S1), we also computed the odds-ratio (“or,” a statistical measure of effect size), in order to assess the strength of the reported associations. Note that our input data, and thus also our predicted associations, likely suffer from undersampling and probably also from systematic biases in the sampling. Both effects are difficult to quantify, but are likely present due to variable choices of PCR primers (information about primers is often not available in the sequence records), and also due to experimental biases in DNA extraction protocols. However, while such biases can likely suppress the detection of certain lineages, it is less likely that they generate false-positive associations at the level of specificity that we observe here (see Supplemental Fig. S3, and see also the randomizations described above). We also noted that, overall, larger samples contribute more co-occurrence associations than smaller samples, as expected. We quantified this in two ways: by stratifying the input data by sample size, and by randomly down-sampling the larger environmental samples (these often focus on the mammalian gut). The results of both tests are summarized in Supplemental Figure S7; reassuringly, we observe that entirely removing gut-related samples through keyword searches, while lowering the number of association clusters, still supports the quantitative conclusion that we report in Figures 4 and 5 (see Supplemental Fig. S10).
Network inference
Based on the co-occurrence analysis results, we constructed networks of coexisting microbes for different levels of OTU definitions. For this, the FDR cutoff for each individual edge in the network was 0.001. In order to obtain a simplified view on the results and to identify cohesive modules of coexisting microbial lineages, we clustered our networks using the Markov cluster algorithm (MCL algorithm; http://micans.org/mcl) (Enright et al. 2002). This clustering was performed using as the similarity metric (i.e., edge weights) the normalized co-occurrence similarity between OTUs, defined here as the Jaccard similarity coefficient (i.e., cooc_count/[(otu1_count + otu2_count) − cooc_count]). We set MCL's “inflation” parameter to 2.0 when running the algorithm. All network images were generated using custom Python scripting and the Python module “NetworkX” (http://networkx.lanl.gov), which provides an interface to the “Graphviz” graph visualization software (http://www.graphviz.org).
Cluster annotation
To annotate clusters in the co-occurrence network with environmental information, we relied on the controlled vocabulary maintained by the Environment Ontology project (EnvO, version 1.51; http://environmentontology.org). In a first step, we assigned EnvO keywords to each OTU in the network; to do so, we scanned all words in the “isolation_source” field from each OTU and assigned ontology terms to that OTU based on exact matches. For many of its terms, EnvO also provides “synonyms”; for cases in which a term could not be matched directly, we also allowed matches via these synonyms, but only for synonyms of the categories “EXACT” or “NARROW” (omitting the categories “RELATED” and “BROAD”). The Fisher's exact test then allowed us to assign significantly over-represented keywords (FDR = 0.01; P-value adjusted for multiple testing using the Benjamini and Hochberg procedure) for each given cluster or subnetwork, compared to the background frequency of these terms in the entire network.
Comparative genomics
First, we mapped available, completely sequenced genomes to our OTUs, for various levels of OTU definitions. For this, we extracted the 16S rRNA genes predicted for 881 complete genomes contained in the RefSeq database (RefSeq 35, 05-13-2009), requiring a minimum length of 700 bp. We then compared these sequences against representative 16S sequences from each OTU, using BLAST with the following parameters: “-a 2 -m 8 -p blastn -v 1000 -b 1000 -r 2 -q -3 -G 5 -E 2 -e 0.01.” For genomes that are annotated with more than one predicted 16S rRNA gene, we retained the longest copy. For the mapping, we then ranked all sequence matches by bit-score (best score first) and, parsing through the list, assigned each genome to the best-matching OTU (skipping those that were already previously assigned to another genome). In addition, we required that the alignment length for the BLAST hit was at least 800 bp and that the sequence identity of the match was 97% or greater.
We then analyzed co-occurring OTUs by comparing their mapped genomes, using several characteristics: genome size, GC content, and relative coverage of KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways (Kanehisa et al. 2008). To compute genome size ratios, we used the total DNA length of the non-redundant chromosomes and plasmids, expressed in nucleotides; to compute GC content ratios, we used the predetermined values for the complete genomes available at http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi. In order to compare genomes in terms of their encoded functions, we assessed the relative coverage of pathways as annotated at KEGG, using the KEGG API (http://www.genome.jp/kegg/soap). We computed normalized vectors describing the relative pathway coverage among all annotated genes of a given genome and then compared these vectors by computing their Euclidean distance. In order to exclude potential artifacts arising from occasional annotation errors in KEGG, we repeated this analysis with two additional, independent systems of functional genome annotation, retrieving essentially the same results (Supplemental Fig. S4).
To test for the statistical independence of our observations made for a given distance measure, against another measure (usually against phylogenetic distance) (Fig. 4E–G), we first learned the dependency between the two measures based on randomly selected pairs of genomes (blue dots). This dependency was then described using a running median (blue lines in Fig. 4). Next, we assessed the data of interest (i.e., pairs of co-occurring genomes) by computing for each data point its vertical distance to the (blue) running median, divided by that median itself. This measure has been termed “relative distance to median” (“dm”; see, for example, Newman et al. 2006); it permits us to compare data at a given, fixed setting of a second, potentially confounding variable. From this, we generated a distribution of normalized distance values, which we compared to the corresponding random background distributions, using the non-parametric Kolmogorov-Smirnov test.
Data availability
Raw input data, as well as all computed results of this study (including sequence data, operational taxonomic units, co-occurrence statistics, network clustering, and genome mapping) are available online at http://mblnx-kallisto.uzh.ch:8888/microbial_coexistence/. In addition, a zoomable and clickable version of the network in Figure 2C is available as Supplemental Figure S12, which can be downloaded from the Supplemental materials.
Acknowledgments
This work was funded by the Swiss National Science Foundation and by the University of Zurich through its Research Priority Program in Systems Biology and Functional Genomics. We thank Phil Hugenholtz and Todd DeSantis for help with the Greengenes database, and Wolf-Dietrich Hardt for insightful comments and criticism.
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.104521.109.
References
- Ahmed N 2009. A flood of microbial genomes—do we need more? PLoS One 4: e5831 doi: 10.1371/journal.pone.0005831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso C, Warnecke F, Amann R, Pernthaler J 2007. High local and global diversity of Flavobacteria in marine plankton. Environ Microbiol 9: 1253–1266 [DOI] [PubMed] [Google Scholar]
- Angly FE, Willner D, Prieto-Davo A, Edwards RA, Schmieder R, Vega-Thurber R, Antonopoulos DA, Barott K, Cottrell MT, Desnues C, et al. 2009. The GAAS metagenomic tool and its estimations of viral and microbial average genome size in four major biomes. PLoS Comput Biol 5: e1000593 doi: 10.1371/journal.pcbi.1000593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baati H, Guermazi S, Amdouni R, Gharsallah N, Sghir A, Ammar E 2008. Prokaryotic diversity of a Tunisian multipond solar saltern. Extremophiles 12: 505–518 [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN 2004. Network biology: Understanding the cell's functional organization. Nat Rev Genet 5: 101–113 [DOI] [PubMed] [Google Scholar]
- Barton HA, Taylor NM, Lubbers BR, Pemberton AC 2006. DNA extraction from low-biomass carbonate rock: An improved method with reduced contamination and the low-biomass contaminant database. J Microbiol Methods 66: 21–31 [DOI] [PubMed] [Google Scholar]
- Bell T, Ager D, Song JI, Newman JA, Thompson IP, Lilley AK, van der Gast CJ 2005. Larger islands house more bacterial taxa. Science 308: 1884. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, Amann R, Jorgensen BB, Witte U, Pfannkuche O 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407: 623–626 [DOI] [PubMed] [Google Scholar]
- Boucher DH 1985. The biology of mutualism: Ecology and evolution Oxford University Press, New York [Google Scholar]
- Brauman A, Kane MD, Labat M, Breznak JA 1992. Genesis of acetate and methane by gut bacteria of nutritionally diverse termites. Science 257: 1384–1387 [DOI] [PubMed] [Google Scholar]
- Caldwell SL, Laidler JR, Brewer EA, Eberly JO, Sandborgh SC, Colwell FS 2008. Anaerobic oxidation of methane: Mechanisms, bioenergetics, and the ecology of associated microorganisms. Environ Sci Technol 42: 6791–6799 [DOI] [PubMed] [Google Scholar]
- Chain PS, Grafham DV, Fulton RS, Fitzgerald MG, Hostetler J, Muzny D, Ali J, Birren B, Bruce DC, Buhay C, et al. 2009. Genomics. Genome project standards in a new era of sequencing. Science 326: 236–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, et al. 2009. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37: D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R 2009. Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bary A 1879. Die Erscheinung der Symbiose Verlag Karl J. Trubner, Strassbourg [Google Scholar]
- Desantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit R, Bouvier T 2006. “Everything is everywhere, but, the environment selects”; what did Baas Becking and Beijerinck really say? Environ Microbiol 8: 755–758 [DOI] [PubMed] [Google Scholar]
- Dubilier N, Mulders C, Ferdelman T, de Beer D, Pernthaler A, Klein M, Wagner M, Erseus C, Thiermann F, Krieger J, et al. 2001. Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm. Nature 411: 298–302 [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30: 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferriere R, Gauduchon M, Bronstein JL 2007. Evolution and persistence of obligate mutualists and exploiters: Competition for partners and evolutionary immunization. Ecol Lett 10: 115–126 [DOI] [PubMed] [Google Scholar]
- Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. 2008a. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 26: 541–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D, Garrity GM, Sansone SA, Sterk P, Gray T, Kyrpides N, Hirschman L, Glockner FO, Kottmann R, Angiuoli S, et al. 2008b. Meeting report: The fifth Genomic Standards Consortium (GSC) workshop. OMICS 12: 109–113 [DOI] [PubMed] [Google Scholar]
- Finlay BJ 2002. Global dispersal of free-living microbial eukaryote species. Science 296: 1061–1063 [DOI] [PubMed] [Google Scholar]
- Forterre P, Gribaldo S, Brochier-Armanet C 2009. Happy together: Genomic insights into the unique Nanoarchaeum/Ignicoccus association. J Biol 8: 7 doi: 10.1186/jbiol110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox GE, Stackebrandt E, Hespell RB, Gibson J, Maniloff J, Dyer TA, Wolfe RS, Balch WE, Tanner RS, Magrum LJ, et al. 1980. The phylogeny of prokaryotes. Science 209: 457–463 [DOI] [PubMed] [Google Scholar]
- Freilich S, Kreimer A, Meilijson I, Gophna U, Sharan R, Ruppin E 2010. The large-scale organization of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Res doi: 10.1093/nar/gkq118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA 2009. Microbial community structure and its functional implications. Nature 459: 193–199 [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Steele JA 2008. Community structure of marine bacterioplankton: Patterns, networks, and relationships to function. Aquat Microb Ecol 53: 69–81 [Google Scholar]
- Garcia-Pichel F, Nubel U, Muyzer G 1998. The phylogeny of unicellular, extremely halotolerant cyanobacteria. Arch Microbiol 169: 469–482 [DOI] [PubMed] [Google Scholar]
- Ghosh A, Dey N, Bera A, Tiwari A, Sathyaniranjan K, Chakrabarti K, Chattopadhyay D 2010. Culture independent molecular analysis of bacterial communities in the mangrove sediment of Sundarban, India. Saline Systems 6: 1 doi: 10.1186/1746-1448-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs KA, Urbanowski ML, Greenberg EP 2008. Genetic determinants of self identity and social recognition in bacteria. Science 321: 256–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Britschgi TB, Moyer CL, Field KG 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345: 60–63 [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M, et al. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242–1245 [DOI] [PubMed] [Google Scholar]
- Goto T, Yamashita A, Hirakawa H, Matsutani M, Todo K, Ohshima K, Toh H, Miyamoto K, Kuhara S, Hattori M, et al. 2008. Complete genome sequence of Finegoldia magna, an anaerobic opportunistic pathogen. DNA Res 15: 39–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Knight R 2009. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res 19: 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S, Rainey P, Haagensen J, Molin S 2007. Evolution of species interactions in a biofilm community. Nature 445: 533–536 [DOI] [PubMed] [Google Scholar]
- Harcombe W 2010. Novel cooperation experimentally evolved between species. Evolution doi: 10.1111/j.1558-5646.2010.00959.x [DOI] [PubMed] [Google Scholar]
- Harrison BK, Zhang H, Berelson W, Orphan VJ 2009. Variations in archaeal and bacterial diversity associated with the sulfate-methane transition zone in continental margin sediments (Santa Barbara Basin, California). Appl Environ Microbiol 75: 1487–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey PH, Colwell RK, Silvertown JW, May RM 1983. Null models in ecology. Annu Rev Ecol Syst 14: 189–211 [Google Scholar]
- Heijs SK, Damste JS, Forney LJ 2005. Characterization of a deep-sea microbial mat from an active cold seep at the Milano mud volcano in the Eastern Mediterranean Sea. FEMS Microbiol Ecol 54: 47–56 [DOI] [PubMed] [Google Scholar]
- Hibbing ME, Fuqua C, Parsek MR, Peterson SB 2010. Bacterial competition: Surviving and thriving in the microbial jungle. Nat Rev Microbiol 8: 15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner-Devine MC, Silver JM, Leibold MA, Bohannan BJ, Colwell RK, Fuhrman JA, Green JL, Kuske CR, Martiny JB, Muyzer G, et al. 2007. A comparison of taxon co-occurrence patterns for macro- and microorganisms. Ecology 88: 1345–1353 [DOI] [PubMed] [Google Scholar]
- Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO 2002. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 417: 63–67 [DOI] [PubMed] [Google Scholar]
- Huber T, Faulkner G, Hugenholtz P 2004. Bellerophon: A program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20: 2317–2319 [DOI] [PubMed] [Google Scholar]
- Hubert C, Loy A, Nickel M, Arnosti C, Baranyi C, Bruchert V, Ferdelman T, Finster K, Christensen FM, Rosa de Rezende J, et al. 2009. A constant flux of diverse thermophilic bacteria into the cold Arctic seabed. Science 325: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Inagaki F, Nunoura T, Nakagawa S, Teske A, Lever M, Lauer A, Suzuki M, Takai K, Delwiche M, Colwell FS, et al. 2006. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc Natl Acad Sci 103: 2815–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenbarger TA, Finney M, Rios-Velazquez C, Handelsman J, Ruvkun G 2008. Miniprimer PCR, a new lens for viewing the microbial world. Appl Environ Microbiol 74: 840–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone RA, Bshary R 2008. Mutualism, market effects and partner control. J Evol Biol 21: 879–888 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittel K, Boetius A 2009. Anaerobic oxidation of methane: Progress with an unknown process. Annu Rev Microbiol 63: 311–334 [DOI] [PubMed] [Google Scholar]
- Kolsto AB, Tourasse NJ, Okstad OA 2009. What sets Bacillus anthracis apart from other Bacillus species? Annu Rev Microbiol 63: 451–476 [DOI] [PubMed] [Google Scholar]
- Kolter R, Greenberg EP 2006. Microbial sciences: The superficial life of microbes. Nature 441: 300–302 [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM 2004. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci 101: 3160–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottmann R, Kostadinov I, Duhaime MB, Buttigieg PL, Yilmaz P, Hankeln W, Waldmann J, Glockner FO 2010. Megx.net: Integrated database resource for marine ecological genomics. Nucleic Acids Res 38: D391–D395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, He S, Warnecke F, Peterson SB, Garcia Martin H, Haynes M, Ivanova N, Blackall LL, Breitbart M, Rohwer F, et al. 2008a. A bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res 18: 293–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Raes J, Harris JK, Spear JR, Walker JJ, Ivanova N, von Mering C, Bebout BM, Pace NR, Bork P, et al. 2008b. Millimeter-scale genetic gradients and community-level molecular convergence in a hypersaline microbial mat. Mol Syst Biol 4: 198 doi: 10.1038/msb.2008.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci 82: 6955–6959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenheder S, Prosser JI 2008. Resource availability influences the diversity of a functional group of heterotrophic soil bacteria. Environ Microbiol 10: 2245–2256 [DOI] [PubMed] [Google Scholar]
- Langenheder S, Lindstrom ES, Tranvik LJ 2006. Structure and function of bacterial communities emerging from different sources under identical conditions. Appl Environ Microbiol 72: 212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Harris JK, Wilcox J, Spear JR, Miller SR, Bebout BM, Maresca JA, Bryant DA, Sogin ML, Pace NR 2006. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl Environ Microbiol 72: 3685–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI 2008. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat Rev Microbiol 6: 776–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Wang P 2008. [Bacterial and archaeal diversity in surface sediment from the south slope of the South China Sea]. Wei Sheng Wu Xue Bao 48: 323–329 [PubMed] [Google Scholar]
- Li T, Wang P, Wang PX 2008. Microbial diversity in surface sediments of the Xisha Trough, the South China Sea. Acta Ecologica Sinica 28: 1166–1173 [Google Scholar]
- Lloyd KG, Lapham L, Teske A 2006. An anaerobic methane-oxidizing community of ANME-1b archaea in hypersaline Gulf of Mexico sediments. Appl Environ Microbiol 72: 7218–7230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Ivanova NN, Szeto E, Palaniappan K, Chu K, Dalevi D, Chen IM, Grechkin Y, Dubchak I, Anderson I, et al. 2008. IMG/M: A data management and analysis system for metagenomes. Nucleic Acids Res 36: D534–D538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, et al. 2006. Microbial biogeography: Putting microorganisms on the map. Nat Rev Microbiol 4: 102–112 [DOI] [PubMed] [Google Scholar]
- McHardy AC, Rigoutsos I 2007. What's in the mix: Phylogenetic classification of metagenome sequence samples. Curr Opin Microbiol 10: 499–503 [DOI] [PubMed] [Google Scholar]
- Min KR, Rickard AH 2009. Coaggregation by the freshwater bacterium Sphingomonas natatoria alters dual-species biofilm formation. Appl Environ Microbiol 75: 3987–3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet 17: 589–596 [DOI] [PubMed] [Google Scholar]
- Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER 2008. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl Environ Microbiol 74: 4530–4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrazek J 2009. Phylogenetic signals in DNA composition: Limitations and prospects. Mol Biol Evol 26: 1163–1169 [DOI] [PubMed] [Google Scholar]
- Muyzer G, Teske A, Wirsen CO, Jannasch HW 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol 164: 165–172 [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR 2009. Infernal 1.0: Inference of RNA alignments. Bioinformatics 25: 1335–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS 2006. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846 [DOI] [PubMed] [Google Scholar]
- Newton RJ, Jones SE, Helmus MR, McMahon KD 2007. Phylogenetic ecology of the freshwater Actinobacteria acI lineage. Appl Environ Microbiol 73: 7169–7176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Bassler BL 2009. Bacterial quorum-sensing network architectures. Annu Rev Genet 43: 197–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The NIH HMP Working Group 2009. The NIH Human Microbiome Project. Genome Res 19: 2317–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson AI, Koskiniemi S, Eriksson S, Kugelberg E, Hinton JC, Andersson DI 2005. Bacterial genome size reduction by experimental evolution. Proc Natl Acad Sci 102: 12112–12116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphan VJ 2009. Methods for unveiling cryptic microbial partnerships in nature. Curr Opin Microbiol 12: 231–237 [DOI] [PubMed] [Google Scholar]
- Overmann J, Schubert K 2002. Phototrophic consortia: Model systems for symbiotic interrelations between prokaryotes. Arch Microbiol 177: 201–208 [DOI] [PubMed] [Google Scholar]
- Palmer TM, Stanton ML, Young TP 2003. Competition and coexistence: Exploring mechanisms that restrict and maintain diversity within mutualist guilds. Am Nat 162: S63–S79 [DOI] [PubMed] [Google Scholar]
- Paracer S, Ahmadjian V 2000. Symbiosis: An introduction to biological associations Oxford University Press, New York [Google Scholar]
- Parsek MR, Greenberg EP 2005. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol 13: 27–33 [DOI] [PubMed] [Google Scholar]
- Pedros-Alio C 2006. Marine microbial diversity: Can it be determined? Trends Microbiol 14: 257–263 [DOI] [PubMed] [Google Scholar]
- Piccini C, Conde D, Alonso C, Sommaruga R, Pernthaler J 2006. Blooms of single bacterial species in a coastal lagoon of the southwestern Atlantic Ocean. Appl Environ Microbiol 72: 6560–6568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO 2007. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Bork P 2008. Molecular eco-systems biology: Towards an understanding of community function. Nat Rev Microbiol 6: 693–699 [DOI] [PubMed] [Google Scholar]
- Raes J, Korbel JO, Lercher MJ, von Mering C, Bork P 2007. Prediction of effective genome size in metagenomic samples. Genome Biol 8: R10 doi: 10.1186/gb-2007-8-1-r10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranea JA, Buchan DW, Thornton JM, Orengo CA 2004. Evolution of protein superfamilies and bacterial genome size. J Mol Biol 336: 871–887 [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A 2000. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, Arellano A, Coleman M, Hauser L, Hess WR, et al. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424: 1042–1047 [DOI] [PubMed] [Google Scholar]
- Ruan Q, Dutta D, Schwalbach MS, Steele JA, Fuhrman JA, Sun F 2006. Local similarity analysis reveals unique associations among marine bacterioplankton species and environmental factors. Bioinformatics 22: 2532–2538 [DOI] [PubMed] [Google Scholar]
- Ruehland C, Blazejak A, Lott C, Loy A, Erseus C, Dubilier N 2008. Multiple bacterial symbionts in two species of co-occurring gutless oligochaete worms from Mediterranean sea grass sediments. Environ Microbiol 10: 3404–3416 [DOI] [PubMed] [Google Scholar]
- Ryan RP, Dow JM 2008. Diffusible signals and interspecies communication in bacteria. Microbiology 154: 1845–1858 [DOI] [PubMed] [Google Scholar]
- Saffo MB 1993. Coming to terms with a field: Words and concepts in symbiosis. Symbiosis 14: 17–31 [Google Scholar]
- Sahl JW, Pace NR, Spear JR 2008. Comparative molecular analysis of endoevaporitic microbial communities. Appl Environ Microbiol 74: 6444–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri R, Kravitz SA, Smarr L, Gilna P, Frazier M 2007. CAMERA: A community resource for metagenomics. PLoS Biol 5: e75 doi: 10.1371/journal.pbio.0050075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank EA, Kolter R 2009. New developments in microbial interspecies signaling. Curr Opin Microbiol 12: 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith VH 2007. Microbial diversity–productivity relationships in aquatic ecosystems. FEMS Microbiol Ecol 62: 181–186 [DOI] [PubMed] [Google Scholar]
- Sorensen KB, Canfield DE, Teske AP, Oren A 2005. Community composition of a hypersaline endoevaporitic microbial mat. Appl Environ Microbiol 71: 7352–7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JT, Konopka A 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol 39: 321–346 [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Komai T, Hanada S, Tamaki S, Tanabe S, Miyachi Y, Uchiyama M, Nakazawa T, Kimura K, Kamagata Y 2009. Bacterial and archaeal 16S rRNA genes in Late Pleistocene to Holocene muddy sediments from the Kanto Plain of Japan. Geomicrobiol J 26: 104–118 [Google Scholar]
- Tanner MA, Goebel BM, Dojka MA, Pace NR 1998. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol 64: 3110–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CS, Ramsey ME, Wang YL, Frost AM, Jun E, Durham T 2009. Minimal genomes, maximal productivity: Comparative genomics of the photosystem and light-harvesting complexes in the marine cyanobacterium, Prochlorococcus. Photosynth Res 101: 1–19 [DOI] [PubMed] [Google Scholar]
- Tokura M, Ohkuma M, Kudo T 2000. Molecular phylogeny of methanogens associated with flagellated protists in the gut and with the gut epithelium of termites. FEMS Microbiol Ecol 33: 233–240 [DOI] [PubMed] [Google Scholar]
- Tringe SG, Hugenholtz P 2008. A renaissance for the pioneering 16S rRNA gene. Curr Opin Microbiol 11: 442–446 [DOI] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43 [DOI] [PubMed] [Google Scholar]
- van Nimwegen E 2003. Scaling laws in the functional content of genomes. Trends Genet 19: 479–484 [DOI] [PubMed] [Google Scholar]
- Vautrin E, Vavre F 2009. Interactions between vertically transmitted symbionts: Cooperation or conflict? Trends Microbiol 17: 95–99 [DOI] [PubMed] [Google Scholar]
- Vilas-Boas GT, Peruca AP, Arantes OM 2007. Biology and taxonomy of Bacillus cereus, Bacillus anthracis, and Bacillus thuringiensis. Can J Microbiol 53: 673–687 [DOI] [PubMed] [Google Scholar]
- Visick KL, Fuqua C 2005. Decoding microbial chatter: Cell–cell communication in bacteria. J Bacteriol 187: 5507–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Mering C, Hugenholtz P, Raes J, Tringe SG, Doerks T, Jensen LJ, Ward N, Bork P 2007. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science 315: 1126–1130 [DOI] [PubMed] [Google Scholar]
- Wanner G, Vogl K, Overmann J 2008. Ultrastructural characterization of the prokaryotic symbiosis in “Chlorochromatium aggregatum.” J Bacteriol 190: 3721–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DM, Weller R, Bateson MM 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345: 63–65 [DOI] [PubMed] [Google Scholar]
- Waters C, Bassler B 2005. Quorum sensing: Cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21: 319–346 [DOI] [PubMed] [Google Scholar]
- Woyke T, Teeling H, Ivanova N, Huntemann M, Richter M, Gloeckner F, Boffelli D, Anderson I, Barry K, Shapiro H, et al. 2006. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 443: 950–955 [DOI] [PubMed] [Google Scholar]
- Zhang W, Ki JS, Qian PY 2008. Microbial diversity in polluted harbor sediments I: Bacterial community assessment based on four clone libraries of 16S rDNA. Estuarine Coastal Shelf Sci 76: 668–681 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw input data, as well as all computed results of this study (including sequence data, operational taxonomic units, co-occurrence statistics, network clustering, and genome mapping) are available online at http://mblnx-kallisto.uzh.ch:8888/microbial_coexistence/. In addition, a zoomable and clickable version of the network in Figure 2C is available as Supplemental Figure S12, which can be downloaded from the Supplemental materials.