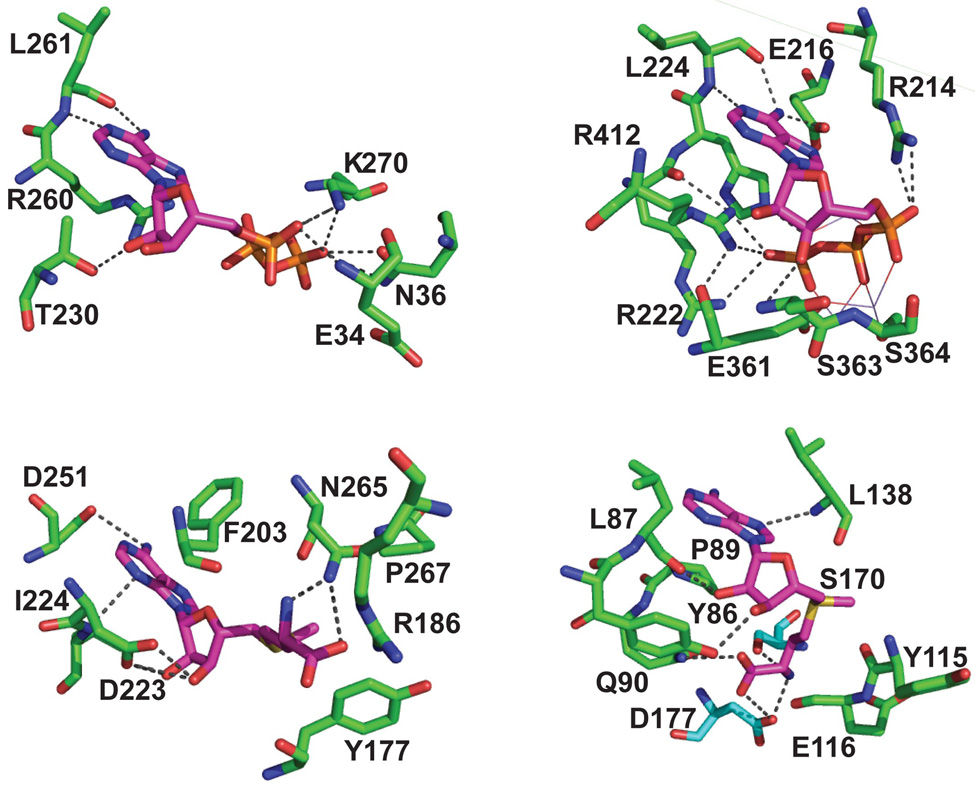
Figure 7.
Active-site structural similarities between AdoMet-dependent methyl transferases and ATP-dependent aminoacyl-tRNA synthetases. (A) Active site of M. jannaschii Trm5 in complex with AdoMet (PDB ID:2ZZN) 9. (B) Active site of Haemophilus influenzae TrmD in complex with AdoMet (PDB ID:1UAK) 4. (C) Active site of the class I E. coli GlnRS in complex with ATP complex (PDB ID:1GTR) 26. (D) Active site of the class II Pyrococcus kodakaraensis AspRS in complex with a U-shaped ATP (PDB ID:1B8A) 51. All structures show the conserved residues that stabilize the specific conformation of the cofactor, which exhibits an extended conformation in (A) and (C) and in a U-shaped conformation in (B) and (D).