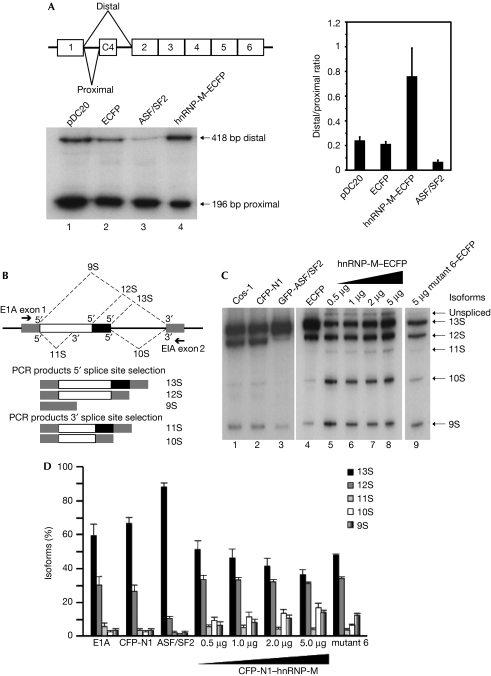
Figure 4.
hnRNP-M is involved in alternative splice site choice through its CDC5L/PLRG1-binding region. (A) Representation of pDC20 mini-gene containing the alternative 3′ splice sites. RT–PCR analysis of HeLa cells co-transfected with pDC20 and either the ECFP-N1 vector (lane 2) or hnRNP-M–ECFP (lane 4). Overexpression of hnRNP-M activates the distal 3′ splice site choice of the chimeric calcitonin/dhfr pre-mRNA. The relative distal-to-proximal 3′ splice site ratio was quantified (right panel). (B) Representation of E1A reporter mini-gene containing the alternative 5′ splice sites. The arrows indicate the positions of the exon primers used for RT–PCR. (C) Analysis of cells co-transfected with E1A and ECFP-N1 vector (lanes 2 and 4) or a gradient of hnRNP-M–ECFP (lanes 5–8) or the mutant 6(248−344aa)–ECFP (lane 9). (D) The relative amounts of the various isoforms were quantified. CDC5L, cell division cycle 5-like; ECFP, enhanced cerculean fluorescent protein; GFP, green fluorescent protein; hnRNP, heterogeneous nuclear ribonucleoprotein; PLRG1, pleiotropic regulator 1; RT–PCR, reverse transcription–PCR.