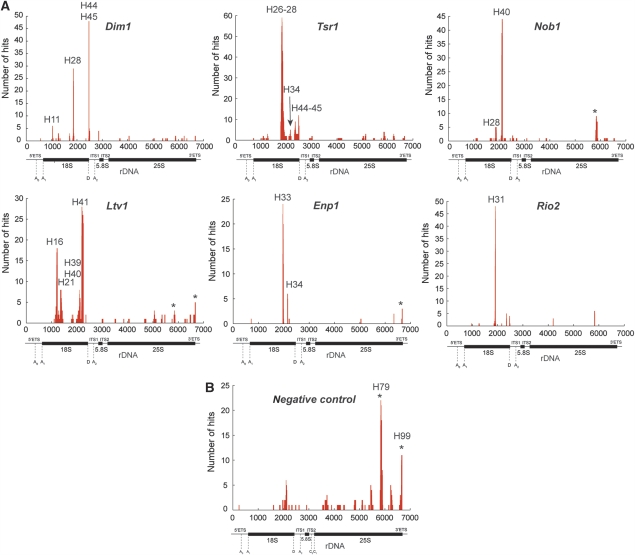
Figure 1.
Overview of CRAC results. Shown are the results from independent CRAC experiments performed on pre-40S-associated proteins (A). Results from untagged strains are shown in (B). Sequences were aligned to the rDNA reference sequence using blast and plotted using gnuplot. The locations of mature rRNA sequences, spacers and cleavage site are indicated below the x axis. The y axis shows the total number of times each nucleotide within an RNA fragment was mapped to the rDNA sequence. The location of the peaks in the secondary structure of the rRNA (see Figure 2A, B) is indicated with helix (H) numbers. The asterisks indicate frequent contaminants.