Abstract
Many DNA lesions cause pausing of replication forks at lesion sites; thus, generating gaps in the daughter strands that are filled-in by post-replication repair (PRR) pathways. In Saccharomyces cerevisiae, PRR involves translesion synthesis (TLS) mediated by Polη or Polζ, or Rad5-dependent gap filling through a poorly characterized error-free mechanism. We have developed an assay to monitor error-free and mutagenic TLS across single DNA lesions in Schizosaccharomyces pombe. For both main UV photolesions, we have delineated a major error-free pathway mediated by a distinct combination of TLS polymerases. Surprisingly, these TLS pathways require enzymes needed for poly-ubiquitination of proliferating cell nuclear antigen (PCNA) as well as those required for mono-ubiquitination. For pathways that require several TLS polymerases the poly-ubiquitin chains of PCNA may facilitate their recruitment through specific interactions with their multiple ubiquitin-binding motifs. These error-free TLS pathways may at least partially account for the previously described poly-ubiquitination-dependent error-free branch of PRR. This work highlights major differences in the control of lesion tolerance pathways between S. pombe and S. cerevisiae despite the homologous sets of PRR genes these organisms share.
Keywords: post-replicative repair, translesion synthesis, ubiquitin ligase Rad6–Rad18, ubiquitin ligase Rad8Rad5/Ubc13–Mms2, UV photoproducts
Introduction
The DNA damage tolerance pathways, also called post-replication repair (PRR), deal with blocks to replication fork progression caused by DNA lesions. These pathways suppress prolonged stalling of DNA replication, allowing lesions to be bypassed and replication to continue. There are two classes of PRR pathways, translesion synthesis (TLS) and DNA damage avoidance (DA). Although the mechanisms of DA, also referred to as template switching, are largely unknown, they involve the transfer of genetic information between the chromatids. This process is thought to take advantage of the replicated undamaged sister chromatid through a template-switching mechanism and is thus deemed to be accurate. In contrast, TLS pathways involve the transient recruitment of specialized DNA polymerases capable of reading through damaged bases, an intrinsically error-prone process that causes mutations in vivo. In eukaryotes, five specialized polymerases have been identified that perform TLS (Prakash et al, 2005). Four of them, Polη, Polι, Polκ and REV1 belong to the Y family of DNA polymerases (Ohmori et al, 2001). The fifth, Polζ, consists of the catalytic subunit REV3 and the accessory factor REV7 and belongs to the B family (Nelson et al, 1996). In contrast to the overall mutagenic effect of TLS, Polη is the only known polymerase to act as a tumour suppressor in humans because of its capacity to accurately replicate through cyclobutane pyrimidine dimer (CPDs), the major UV-induced lesions (Masutani et al, 2000). Mutations in the Polη gene cause the variant form of xeroderma pigmentosum (XP-V), which has a very high risk of sunlight-induced skin cancer (Lehmann et al, 1975; Johnson et al, 1999; Masutani et al, 1999).
In Saccharomyces cerevisiae, epistasis analysis suggests that the PRR pathway is controlled by two master genes RAD6 and RAD18 and further subdivided into three sub-pathways, two TLS pathways defined by Polη and Polζ and an error-free template-switching pathway. The two crucial proteins, RAD6 and RAD18, have E2-ubiquitin conjugating and E3-ubiquitin ligase activities, respectively. The target in PRR of these ubiquitinating enzymes is proliferating cell nuclear antigen (PCNA), which serves as the processivity factor for the replicative and specialized DNA polymerases. In response to DNA damage in mammals and in yeast, PCNA is mono-ubiquitinated at lysine (K) 164 by RAD6 and RAD18 (RAD6/RAD18 complex) (Hoege et al, 2002; Kannouche et al, 2004; Watanabe et al, 2004). In S. cerevisiae, it has been shown that the mono-ubiquitinated form of PCNA promotes UV- and MMS-induced mutagenesis (Stelter and Ulrich, 2003). In higher eukaryotes the importance of PCNA mono-ubiquitination for the TLS pathway has been shown by the fact that specialized TLS polymerases have a stronger affinity for the mono-ubiquitinated form of PCNA (Kannouche et al, 2004; Watanabe et al, 2004) by virtue of ubiquitin-binding motifs found in all four of the Y-family polymerases (Bienko et al, 2005; Guo et al, 2006; Plosky et al, 2006). In vitro studies using yeast enzymes have shown the ability of Polη or Rev1 to carry out TLS stimulated by ubiquitinated PCNA (Garg and Burgers, 2005; Zhuang et al, 2008). Mono-ubiquitinated PCNA can subsequently be poly-ubiquitinated through a K63 linkage by the E2 Ub-conjugating enzyme, UBC13/MMS2, and the E3 Ub ligase RAD5 (Parker and Ulrich, 2009). According to current models, the poly-ubiquinated form of PCNA favours the error-free bypass of DNA lesions by means of DA strategies (Hoege et al, 2002; Moldovan et al, 2007).
In Schizosaccharomyces pombe, Rhp6 and Rhp18, the homologs of the S. cerevisiae RAD6 and RAD18, and Rad8 (the RAD5 homolog) associated with Mms2–Ubc13 are responsible for the mono- and poly-ubiquitination of PCNA on lysine 164, respectively (Frampton et al, 2006). Although absent in S. cerevisiae, Polκ (dinB) is one of the TLS polymerases in the fission yeast together with Polη (eso1), Polζ (rev3 and rev7) and Rev1 (rev1). To gain insight into the mechanisms of TLS pathways and their control, we designed a molecular tool that allows TLS to be monitored in fission yeast. In this study, we analyse the genetics of TLS across the two most common UV-induced lesions, namely, the TT-CPD and TT(6-4) photoproducts. For both the lesions, we find that the major TLS pathway is error-free. For TT-CPD, the major TLS pathway depends on Polη and to a lesser extent on Polκ, whereas TLS across TT(6-4) requires Polη, Polζ and Rev1. Surprisingly, TLS past either lesion is largely dependent on the ubiquitin ligase complex (Rad8Rad5/Ubc13–Mms2), that is, responsible for poly-ubiquitination of PCNA at residue K164.
Results
Development of a TLS assay in S. pombe
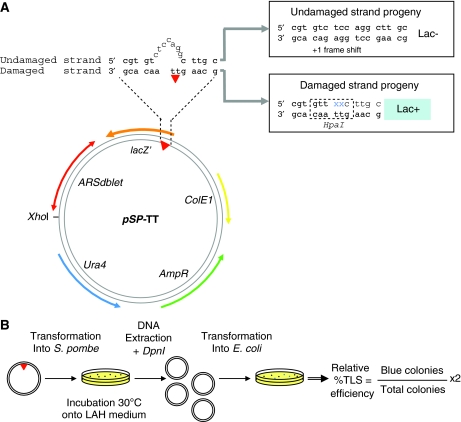
Principle of the assay. The plasmids used in this work contains a defined replication origin and a single lesion located within a short sequence heterology that allows the replication pattern of the two strands to be analysed independently (Figure 1A). In Escherichia coli, when a plasmid containing a single replication-blocking lesion in one strand is replicated, functional uncoupling of the replication machinery occurs at the lesion site (Pages and Fuchs, 2003). Evidence for replicative uncoupling has also been obtained in chromosomes of S. cerevisiae after UV irradiation (Lopes et al, 2006). Indeed, the polymerase that replicates the lesion-containing strand is transiently stalled at the lesion site whereas replication of the undamaged strand proceeds unperturbed as shown both in vivo (Pages and Fuchs, 2003) and in vitro (Higuchi et al, 2003; McInerney and O'Donnell, 2004). The undamaged strand replicates with the same kinetics as either of the strands in an undamaged plasmid (Pages and Fuchs, 2003). Such plasmids are ideal tools to monitor TLS in vivo as they deliver the lesion in the context of a genuine replication fork. The present plasmid assay accurately measures the efficiency of replication across a single lesion compared with the efficiency of replication of the non-damaged strand that acts as an internal standard. Indeed, in the first replication cycle, the damage-containing strand will suffer from a delay that reflects the intrinsic difficulty to bypass the lesion under analysis, whereas the undamaged strand replicates with unmodified kinetics. In the following cycles, the daughter strand that results from the TLS event will now undergo cycles of amplification with the same kinetics as the undamaged strand. After many cycles of replication in S. pombe, the replicated plasmid mixture is extracted, digested with DpnI to eliminate non-replicated DNA and transformed into E. coli for determination of the relative TLS efficiency (RTE) (see below).
Figure 1.
Outline of the TLS assay in S. pombe. (A) Scheme of the pSP-TT plasmid construct. The shuttle plasmid contains the ColE1 origin, the ampR resistance cassette, the bi-directional S. pombe ARSDblet replication origin and the URA4 marker for propagation and selection in E. coli and S. pombe, respectively. The single TT lesion, either a cyclobutane (CPD) or 6-4 photoproduct, is located at the beginning of the lacZ′ reporter gene opposite a short sequence heterology. Replication of the lesion-containing strand (DAMAGED strand) by TLS yields a functional lacZ′ gene whatever nucleotides are inserted opposite the TT lesion whereas the NON-DAMAGED strand carries a +1 frameshift that prevents lacZ expression. (B) Flow chart of the experimental design. The covalently closed circular plasmid carrying the single lesion is introduced into S. pombe cells by transformation. After a 72 h period of replication in the yeast cells, the plasmid is extracted, and the DpnI-resistant plasmid replication products are transformed into E. coli cells and plated on ampicillin X-gal plates. The RTE across a given lesion (TLS %) is defined as the proportion of blue colonies (LacZ+) divided by the total number of colonies multiplied by two (see Results paragraph). The TLS events (blue colonies) are further analysed by sequencing to determine the respective proportions of error-free and mutagenic TLS events.
Specificity of the vector used in S. pombe assay.To develop such an assay, the choice of a proper replication origin that is able to stably maintain a plasmid in S. pombe was critical. We chose the sequence ARSdblet, a bi-directional autonomous replication sequence, known to maintain plasmids as monomers during many generations in S. pombe (Brun et al, 1995). Avoiding multimerisation improves the transmission of the plasmid through mitosis and increases the efficiency of subsequent plasmid analysis in E. coli. The plasmid construct (pSP) also carries the URA4 marker for selection in fission yeast and the ColE1 origin and the ampR cassette for propagation and selection in E. coli.
A single TT lesion, either a CPD or 6–4 photoproduct, is located at the beginning of the lacZ′ reporter gene opposite a short sequence heterology that serves as a genetic strand marker (Figure 1A). To prevent repair of the lesion and of the sequence heterology, the TLS assay was performed in a strain deficient for nucleotide excision repair (NER), the UV dimer endonuclease-mediated second excision repair process (UVR) in S. pombe and mismatch repair (MMR) (named num parental strain). In this strain, genes swi10, uve1 and mlh1 required for NER, UVR and MMR pathways were knocked out, respectively. Replication of the lesion-containing strand by TLS yields a functional lacZ′ gene (blue colony in E. coli) whatever nucleotides are inserted opposite the TT lesion, because the TT lesion is part of an in-frame valine codon (GTT) in the N-terminal region of the lacZ gene that can tolerate any amino-acid substitution. In contrast, replication of the non-damaged strand yields plasmids that carry a +1 frameshift giving rise to a Lac− phenotype (white colony in E. coli). Our assay is not able to detect frameshift TLS events but in many different systems and organisms, frameshifts induced by UV lesions are rare events (Lawrence et al, 1993).
DA events are not scored in our assay, as they would generate Lac− plasmids as a result of the presence of the local sequence heterology and would thus not be monitored as TLS events. This is discussed further below. Note that we do not suggest that this plasmid system provides an accurate reflection of all the events that might occur in genomic DNA when a replication fork encounters DNA damage. Instead we use it as a system to address a specific question, namely, what are the genetic requirements of TLS.
Validation of the TLS assay.
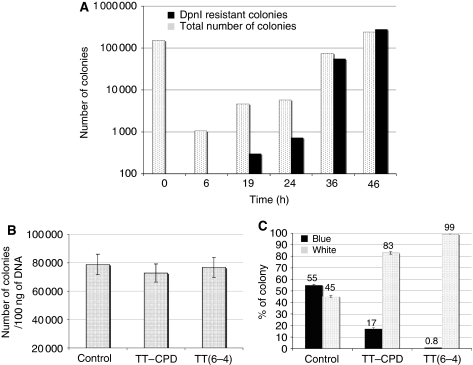
Kinetics of replication of control plasmid construct. First, we monitored the kinetics of replication of the pSP plasmid construct in S. pombe. For this purpose control lesion-free pSP plasmid was transformed into S. pombe cells by electroporation and incubated in selective liquid medium at 30°C. At various times, total DNA was extracted from aliquots of the culture and subjected or not to DpnI digestion. The kinetics of appearance of plasmid DNA replicated in S. pombe cells was assessed by the number of colonies formed in E. coli by the DpnI-resistant plasmid sample extracted from yeast cells. As shown in Figure 2A, colonies formed by the DpnI-resistant plasmid sample appeared at 19 h and increased exponentially until by 46 h they comprised the majority of the population. These data clearly show that the plasmid construct pSP undergoes efficient replication in S. pombe cells. On the basis of these results, we decided to monitor TLS after a period of growth in S. pombe cells of at least 72 h at 30°C.
The two strands of the plasmid are replicated independently in S. pombe. For a given amount of plasmid DNA, we observe the same efficiency of colony formation in S. pombe for unmodified control plasmid as well as for plasmids containing the single TT-CPD or TT(6-4) lesions (Figure 2B). Our hypothesis to explain the observed results is that even if the replication of one strand is blocked by the presence of a lesion, the other, undamaged, strand gets fully replicated and amplified and will thus give rise to colonies with the same efficiency as the control, lesion-free, construct (Pages and Fuchs, 2003). This observation holds in all strains tested and again strongly supports the notion that the present plasmid system does not record DA events (see Discussion below).
Determination of the RTE. We next checked the ability of our assay to monitor the synthesis of each strand of double-stranded DNA during replication. For this purpose, we used the lesion-free construct pSP-(CONTROL) and scored Lac+ and Lac− colonies at the 72 h time point. In principle, in the absence of a lesion, the semi-conservative nature of DNA replication should yield equivalent amounts of progeny derived from each strand. In fact, we observed a ≈55/45 ratio of blue/white colonies, a ratio close to the expected 50/50 value (Figure 2C). Sequencing of individual blue and white colonies yielded the lacZ′+ and lacZ′− (+1 frameshift) sequences derived from each strand of the heteroduplex plasmid construct, respectively. We will define the ‘RTE'=2 × (number of blue colonies)/(total number of colonies). The factor 2 is introduced to take into account the replication of the undamaged strand. Thus, for an undamaged control construct, the efficiency of replication of the Lac+ strand is normalized to ≈100%. When the pSP contruct containing a single TT-CPD or TT(6-4) lesion was replicated in S. pombe, the RTE dropped from ≈100% for the lesion-free control to 34 and 1.6%, respectively. This observation is in good agreement with the notion that though a CPD lesion is a moderate replication block, a (6–4) photoproduct is clearly a severe impediment to replication. Interestingly, we did not observe any bias in TLS events whether the lesion was located on the leading or the lagging strand (data not shown). This observation also suggests that replication of the two strands is uncoupled and can be considered as independent events as previously observed in E. coli (Pages and Fuchs, 2003).
Figure 2.
Validation of the TLS assay. (A) Kinetics of replication of a lesion-free pSP plasmid in S. pombe. At various time points, total DNA was extracted from aliquots of the culture and subjected or not to DpnI digestion. We monitor the kinetics of appearance of plasmid DNA replicated in S. pombe cells by assessing the amount of colonies formed in E. coli. The plasmid replicated in S. pombe is represented by DpnI-treated sample (black bar) and the total amount of plasmid extracted (no DpnI treatment: grey bar). At time point 46 h, essentially all plasmids extracted from S. pombe are resistant to DpnI digestion, proving that they have been replicated in the yeast cells. (B) S. pombe transformation efficiency with control, TT-CPD and TT(6-4) plasmids in the parental numWT strain. (C) TLS assay in the parental numWT strain with lesion-free control plasmid (CONTROL) and plasmids carrying a single TT-CPD or TT(6-4) lesion. The relative amounts of white and blue colonies are determined in E. coli after replication in S. pombe during 72 h. The proportions expressed are per cent (%) are indicated on top of each bar. Error bars result from at least three independent experiments. As expected, with the lesion-free control plasmid both strands yield a similar number of plasmid progeny. We have no explanation for the slight bias (55:45) that is observed. With the lesion-containing constructs, the proportion of blue colonies dramatically decreases, reflecting the replication-blocking potential of the lesions (see text).
Therefore, we analysed the replication of the pSP-TT-CPD and the pSP-TT(6-4) plasmids into different genetic backgrounds. The number of colonies recovered after transformation of each S. pombe strain is shown in Supplementary Table S1. For all tested strains, the number of colonies recovered lies within a range comprised approximately between 4 and 9 × 104 colonies.
TLS across TT-CPD
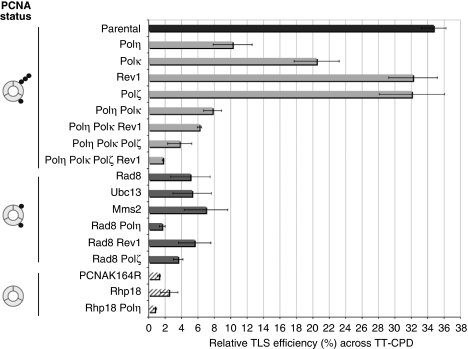
Which DNA polymerases are involved? In the parental strain, the RTE for the TT-CPD lesion is ≈34%, a value that reflects a distinct though moderate replication-blocking capacity when compared with the replication efficiency of lesion-free plasmid (100%) (Figure 3). On inactivation of Polη the TLS efficiency decreased three- to four-fold reaching an RTE value of ≈10%. A major role of Polη in TT-CPD bypass is in good agreement with results from S. cerevisiae and human cells (Gibbs et al, 2005; Hendel et al, 2008). More surprisingly, inactivation of Polκ also led to a two-fold reduction in the extent of TLS. Polη and Polκ appear to control the same pathway as no significant decrease in TLS was observed in the Polη Polκ double mutant compared with the Polη single mutant strain. Sequencing of the progeny plasmids showed that essentially all TLS events are error-free in the parental background. Even in the Polη-deficient strain, >90% of the TLS events are error-free. Given the scarcity of mutagenic bypass events (<3% of TLS events in the wild-type strain), we did not analyse the genetics of the mutagenic pathway for TT-CPD. Inactivation of Polζ or Rev1 had essentially no effect on TLS past the TT-CPD lesion. However, inactivation of Polζ (rev3Δ strain) in the Polη Polκ double mutant significantly reduced the RTE from ≈10 to 4%, suggesting the existence of a minor Polζ-mediated pathway that becomes apparent in the absence of the Polη/Polκ pathway. Taken together, these results suggest that the major TT-CPD bypass pathway is error-free and mediated by Polη together with Polκ. We hypothesize that though in vitro Polη is able to perform both the insertion and extension steps, Polκ frequently contributes to the extension steps in vivo as also found in human cells (Ziv et al, 2009).
Figure 3.
Genetic control of the error-free TLS pathways across TT-CPD in S. pombe. The RTE is monitored in various strains carrying mutations in genes coding for DNA polymerases specialized in TLS or in genes affecting the ubiquitination status of PCNA or combination mutants. For sake of clarity, the strains are grouped with respect to the status of PCNA ubiquitination. Error bars result from at least three independent experiments. For TT-CPD essentially all TLS events (>99%) are error-free in the parental strain. The number of colonies recovered after transformation of each S.pombe strain is shown in Supplementary Table S1.
Role of PCNA ubiquitination. For this purpose, we used strains defective in genes involved in the mono- and poly-ubiquitination of PCNA as well as a PCNA mutant (pcn1-K164R) that cannot be ubiquitinated, (Frampton et al, 2006). TLS past the TT-CPD lesion is fully abolished both in the Δrhp18 and the pcn1-K164R mutant strains thus showing that bypass of the TT-CPD lesion absolutely depends on ubiquitination of PCNA. The residual low level of error-free TLS events (1–2%), that is, observed in pcn1-K164R strain and on simultaneously inactivation of the Polη, Polκ, Polζ and Rev1 polymerases most likely reflects a small contamination of the TT-CPD construct with lesion-free oligonucleotide. Strikingly, in all the three strains that inactivate poly-ubiquitination of PCNA (Δrad8, Δubc13 or Δmms2) whereas leaving mono-ubiquitination intact (Supplementary Figure S1) (Frampton et al, 2006), we observe a reduction of TLS to a level similar to that observed in the Polη/Polκ-deficient strains. This unexpected finding suggests that the major Polη/Polκ pathway requires the Rad8Rad5/Ubc13–Mms2 complex most likely for its role in PCNA poly-ubiquitination. Although no other targets of Rad8Rad5/Ubc13–Mms2 are presently known, we cannot formally exclude the possibility that this ubiquitin ligase complex also targets a protein other than PCNA, that is, essential for TLS.
In the Δrad8 background where PCNA can be mono-ubiquitinated but not poly-ubiquitinated, the residual level of TLS (≈6%) is further reduced to background level (≈2%) when rad30 (Polη) is inactivated, suggesting that there is a small fraction of the Polη/Polκ-mediated TLS that can function with mono-ubiquitinated PCNA. Moreover, we can deduce that the minor Polζ-dependent pathway that becomes clearly apparent in the Polη/Polκ defective background also requires the Rad8Rad5/Ubc13–Mms2 poly-ubiquitination complex (Figure 5A).
TLS across TT(6-4)
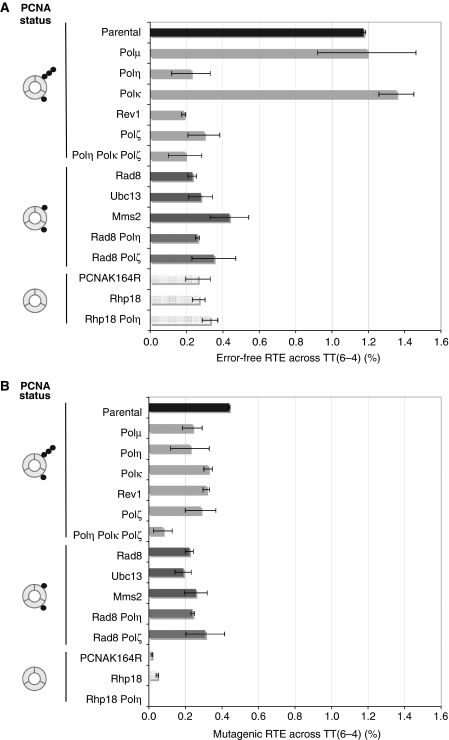
For TT(6-4), the RTE (error-free+mutagenic) (Figure 4) in the parental strain (≈1.6%) is at least 20-fold lower than for TT-CPD illustrating its much stronger replication-blocking capacity (Figure 2C). Analysis of the molecular nature of the TLS events shows that in the parental strain, about three quarters (1.2%) and one quarter (0.4%) of the TLS events are error-free and mutagenic, respectively. Most mutations were either 3′-T to C transitions as in S. cerevisiae (Bresson and Fuchs, 2002) or tandem TT to CC changes (Table 1). In most strains tested, these two types of mutation occur at roughly equal proportion except for Δubc13 and Δrev3 strains, which exhibit a significantly higher proportion of TT → CC changes compared with TT → TC changes (Table 1). We will analyse separately the genetic requirements of error-free and mutagenic TLS pathways across TT(6-4) lesions.
Figure 4.
Genetic control of TLS pathways across TT(6-4) in S. pombe. The RTE is monitored in various strains as described in Figure 3. Error bars result from at least three independent experiments. The number of colonies recovered after transformation of each S.pombe strain is shown in Supplementary Table S1. (A) Error-free TLS pathway across TT(6-4). (B) Mutagenic TLS pathway across TT(6-4).
Table 1. Mutagenesis spectrum in response to TT(6-4) bypass.
| Strain | Number of colonies analysed | Error-free colonies | Mutagenic colonies | Sequence 5′ → 3′ |
|---|---|---|---|---|
| num parental | 33 | 24 | 9 | TT to TC (6) |
| TT to CC (3) | ||||
| num rad30− | 20 | 10 | 10 | TT to TC (5) |
| TT to CC (3) | ||||
| TT to CC+ (2) | ||||
| num dinB− | 31 | 25 | 6 | TT to TC (3) |
| TT to CC (3) | ||||
| num rev1− | 27 | 10 | 17 | TT to TC (11) |
| TT to CC (6) | ||||
| num rev3− | 47 | 23 | 24 | TT to TC (4) |
| TT to CC (19) | ||||
| TT to CC+ (1) | ||||
| num rad8− | 49 | 25 | 24 | TT to TC (12) |
| TT to CC (12) | ||||
| num ubc13− | 47 | 28 | 19 | TT to TC (2) |
| TT to CC (16) | ||||
| TT to AC (1) | ||||
| num mms2− | 23 | 13 | 10 | TT to TC (5) |
| TT to CC (4) | ||||
| TT to AC (1) | ||||
| num K164R | 17 | 16 | 1 | TT to CC (1) |
| num rhp18− | 27 | 23 | 4 | TT to TC (1) |
| TT to CC (3) | ||||
| (+) Additional mutations in the vicinity of the lesion. | ||||
Error-free TLS pathway across the TT(6-4) lesion. Inactivation of either Polη or Polζ or Rev1 led to a four-fold reduction in TLS when compared with the parental strain (Figure 4A). No effect is seen on inactivation of Polκ. These data are consistent with previous in vitro and in vivo studies showing that the bypass of a TT(6-4) lesion often requires the concerted action of two polymerases. In vitro, it has been shown that Polη is able to insert a nucleotide opposite the 3′T of the TT(6-4) photoproduct but unable to extend the mismatched termini while Polζ is specialized for the extension step (Johnson et al, 2001). The precise role of Rev1 is still not known but given its known interactions with Polζ and Polη, it has been suggested that Rev1 acts as a cofactor that mediates the switching between the two polymerases (Friedberg et al, 2005). The residual low level of error-free TLS events (0.2–0.4%) that is observed on inactivation of the Polη/Polζ/Rev1 pathway and also in the pcn1-K164R strain most likely reflects a small contamination of the TT(6-4) construct with lesion-free oligonucleotide. In strains that affect only poly-(Δrad8, Δubc13 or Δmms2) or both mono- and poly-ubiquitination (Δrhp18 or pcn1-K164R) of PCNA, we observe a reduction in TLS similar to that found in the Polη or Polζ-deficient strains suggesting that the Polη/Polζ pathway requires PCNA ubiquitination and the action of the Rad8Rad5/Ubc13–Mms2 complex. In a Polη or Polζ-deficient background, inactivation of rad8 or rph18 did not further reduce the level of TLS. In conclusion, the error-free TLS pathway across TT(6-4) involves the combined action of Polη, Polζ and Rev1. Similarly to TT-CPD bypass it also requires the Rad8Rad5/Ubc13–Mms2 complex most likely for its role in PCNA poly-ubiquitination (Figure 5B).
Figure 5.
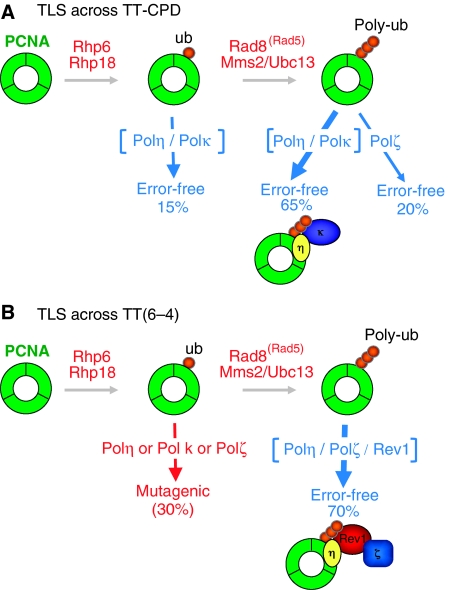
The ‘hanging tool-belt' model: recruitment of multiple TLS polymerases by PCNA poly-ubiquitination in S. pombe. The major TLS pathways across TT-CPD (A) or TT(6-4) (B) lesions in S. pombe are error-free and involve distinct sets of TLS polymerases: Polη and Polκ for TT-CPD, Polη, Rev1 and Polζ for TT(6-4). These pathways strictly depend on ubiquitination at residue K164 of PCNA through the Rad6/Rad18 complex. Moreover, these TLS pathways require a functional Rad8Rad5/Ubc13–Mms2 complex that is known to attach K63-linked poly-ubiquitin chains to mono-ubiquitinated PCNA. We propose that in addition to the known interactions of these polymerases with PCNA (or among themselves), their interaction with the K63-linked poly-ubiquitin chain of PCNA facilitates their assembly into multi-polymerase complexes involved in TLS. In S. pombe, Polη and Polκ possess one UBZ domain, whereas Rev1 contains three UBM domains. Several minor TLS pathways are also inferred from careful analysis of the genetics data.
Mutagenic TLS pathway across TT(6-4). The mutagenic bypass pathway accounts for about one quarter (0.4%) of all TLS events across the TT(6-4) lesion (Figure 4B). Inactivation of each of Polη, or Polκ or Polζ or Rev1 did not yield a significant decrease in the efficiency of mutagenic bypass of the TT(6-4) photoproduct compared to the parental strain. Moreover, in a pol4Δ (Polμ) strain (Gonzalez-Barrera et al, 2005) no reduction in mutagenic TLS efficiency was observed (Figure 4B). However, in a triple polymerase mutant (Polη Polκ Polζ) we see a distinct decrease in the mutagenic bypass efficiency (Figure 4B). In contrast, in S. cerevisiae, mutagenic TLS across TT(6-4), that is, the mis-insertion of a G across the 3′-T, is mediated by Polη followed by Polζ extension (Bresson and Fuchs, 2002).
There was no significant reduction in TLS in any of the three strains that abrogate poly-ubiquitination of PCNA (Δrad8, Δubc13 or Δmms2). However, mutagenic TLS was strongly reduced when mono-ubiquitination of PCNA was also suppressed (Δrhp18 or pcn1-K164R), indicating that this pathway requires mono-ubiquitinated PCNA but not the Rad8Rad5/Ubc13–Mms2 ubiquitin ligase complex. Similarly, in a Δrad30 (Polη) background, though inactivation of rad8 did not reduce TLS, inactivation of rph18 dramatically reduced mutagenic TLS.
In conclusion, mutagenic TLS across TT(6-4) requires the mono-ubiquitinated form of PCNA and is suppressed on simultaneous inactivation of Polη, Polζ and Polκ (Figure 5B).
Discussion
Recently, TLS pathways across specific UV lesions have been analysed in mice and human cells using either a gap-filling assay (Shachar et al, 2009) or an SV40-based double-stranded plasmid DNA replication assay (Yoon et al, 2009). These papers identify the nature of the specific combinations of specialized DNA polymerases involved in these pathways. In this study, in addition to the determination of the specific DNA polymerases involved in the bypass of these lesions in S. pombe, we focus on the control of these TLS pathways by the PRR genes involved in the post-translational modification of PCNA by ubiquitin in S. pombe.
Genetic control of TLS pathways in S. pombe
In the present work, we describe the genetic control of TLS across two common UV-induced lesions, TT-CPD and TT(6-4), in S. pombe. For each lesion, the major TLS pathway is error-free and requires a specific combination of TLS polymerases, namely, Polη/Polκ and Polη/Polζ/Rev1 for TT-CPD and TT(6-4), respectively (Figure 5). As found earlier in S. cerevisiae and in human cells, a point mutation in the replication processivity factor PCNA at position K164, which abolishes Rad6/Rad18-mediated ubiquitination, also eliminates TLS (Hoege et al, 2002; Stelter and Ulrich, 2003; Kannouche et al, 2004). Surprisingly, however, in S. pombe, TLS also requires a functional Rad8Rad5/Ubc13–Mms2 ubiquitin ligase complex. This complex is known to be responsible for the formation of K63-linked poly-ubiquitin chains added onto the K164-linked mono-ubiquitin in PCNA (Parker and Ulrich, 2009; Ye and Rape, 2009). Although we have not formally proven that the observed defect in TLS in Δrad8, Δubc13 or Δmms2 strains solely reflects a deficiency in PCNA poly-ubiquitination, this is, very likely because PCNA is the only known poly-ubiquitination target for the Rad8Rad5, and rad8 and mms2 are epistatic with pcn1-K164R for UV survival (Frampton et al, 2006).
Damage avoidance
Any DA events occurring in our system would not be scored, as they would result in white colonies. However, we do not think that they contribute significantly in our system for the following reasons: (i) DA events, such as template switching or fork regression, require the two sister chromatids to be maintained in close proximity, a structure formed on transient replication fork stalling. As discussed above, the plasmid probes used in the present work are fully unwound by the replicative helicase; thus, not forming the intermediate structures prone to undergo DA events. (ii) If DA did indeed contribute significantly in this system and poly-ubiquitination of PCNA stimulated this process, we would expect that deletion of the genes involved in this process would result in a decrease in the number of white colonies, and therefore a consequent increase in the proportion of blue colonies. In fact, we observed the opposite, a decrease in the proportion of blue colonies.
A possible model for the requirement of poly-ubiquitination of PCNA in TLS
The present results suggest that, in S. pombe, poly-ubiquitination of PCNA is important for TLS pathways involving two specialized DNA polymerases, as exemplified here for the error-free bypass pathways of TT-CPD and TT(6-4) (Figure 5). How may poly-ubiquitination of PCNA facilitate TLS? Although we have no biochemical evidence as yet, it is tempting to speculate that the poly-ubiquitin chains facilitate TLS by assisting the recruitment on PCNA of the various factors involved in lesion bypass. We propose a model referred to as the ‘hanging tool-belt' model in which the coordinated action of two polymerases is facilitated by the concomitant interaction of their UBM or UBZ motifs with distinct ubiquitin moieties in the poly-ubi chain on PCNA. For TT-CPD bypass, Polη and Polκ would thus be recruited, whereas Polη and Polζ would be enlisted through Rev1 for TT(6-4) bypass. Although S. pombe Rev1 contains three UBM motifs (two in other organisms), Polη and Polκ contain one UBZ motif (though Polκ has two in other organisms). Recruitment of proteins through multiple interactions with distinct ubiquitin moeties in K63-linked chains has gained recent experimental evidence (Sato et al, 2009; Walters and Chen, 2009). The two ubiquitin-interacting motifs (UIM1 and UIM2) present in protein Rap80 (receptor-associated protein 80) were shown to interact simultaneously with two neighbouring K63-linked ubiquitin moieties, thereby resulting in recruitment of the BRCA1-Rap80 to K63-linked poly-ubi chains on H2AX in response to double-strand breaks. The assembly of the proteins that will form a complex competent for TLS may thus arise from the sum of various interactions involving direct interaction between the proteins themselves (PCNA–polymerases, polymerases–polymerases) and/or between ubiquitin-binding sites with poly-ubiquitin chains.
Differences between S. pombe and S. cerevisiae in PRR control
The genetic control of TLS past an AP site in S. cerevisiae involves the mono-ubiquitinated form of PCNA as well as the Rad5 protein in a function that does not require its helicase or ubiquitin ligase activities (Pages et al, 2008). The involvement of Rad5 in TLS has also been observed for the bypass of TT(6-4) and G-AAF adducts (Zhang and Lawrence, 2005; Pages et al, 2008). In S. cerevisiae, TLS was not affected in mms2Δ or ubc13Δ strains, suggesting that the role of Rad5 in TLS is distinct from its role as a partner of the E2–E3 ubiquitin complex (Pages et al, 2008). The role of Rad5 in TLS in S. cerevisiae is thus likely to be a ‘structural' role, Rad5 mediating the switch between two specialized DNA polymerases (Pages et al, 2008). In contrast, in S. pombe, error-free TLS across TT-CPD or TT(6-4) is equally affected in all three mutants (rad8Δ or ubc13Δ mms2Δ) that inactivate the E2–E3 complex (Figures 3 and 4), although we have not formally proven that it is the ubiquitin ligase activity of Rad8Rad5 on PCNA that is required for TLS. The notion that Rad5 in S. cerevisiae and Rad8Rad5 in S. pombe have distinct functions in the PRR pathways is also supported by UV survival data. Although in S. cerevisiae, the UV sensitivity of rad5Δ mutants is much more pronounced than that of mms2Δ and ubc13Δ strains (Gangavarapu et al, 2006), the UV sensitivities of rad8Δ, mms2Δ and ubc13Δ strains are similar in S. pombe and the three strains are epistatic (Frampton et al, 2006).
In conclusion, the present work questions the accepted dogma for the role played by poly-ubiquitination of PCNA in post-replication repair and highlights major differences between S. cerevisiae and S. pombe in the control of post-replication repair strategies. These observations warn us against hazardous extrapolation from S. cerevisiae to S. pombe and even more so to human cells even though these organisms share sets of homologous PRR genes.
Materials and methods
General techniques
S. pombe methods and media have been described in Moreno et al (1991).
Strains
The S. pombe strains used in this study are listed in Table 2. Each mutant strain was checked by PCR to verify that it contained the correct genomic alteration, and its phenotype was examined to ensure its consistency with expectation. The polη-deficient strain was made by replacement of the Rad30 domain of Eso1 protein by the KanMX6 cassette. The essential Ctf7 domain of Eso1 is under the control of the nmt promoter. This construct has been checked by Southern blot and no phenotypical differences were observed between wild type and Δrad30 mutant. Rhp6 proved to be unsuitable for experimental use because of its propensity to acquire suppressor mutations. To create the strain pcn1-K164R∷KanMX6, a DNA fragment was generated by overlapping PCR containing pcn1-K164R (Frampton et al, 2006) fused to the KanMX6 cassette flanked with 5′UTR and 3′UTR of the pcn-1 locus. This PCR fragment has been used to replace the wild-type copy of pcn-1 in the S. pombe genome. We confirmed that the strain was UV sensitive and verified that no ubiquitinated form of PCNA was detected by western blot (data not shown; Supplementary Figure S1B).
Table 2. Schizosaccharomyces pombe strains used in this study.
| Strain | Disruption | Origin |
|---|---|---|
| 972h wild type | None | |
| SC323 pcn1-K164R | pcn1-K164R∷kanMX6 | This study |
| SC248 Δrhp18 | rhp18∷hph | This study |
| SC210 Δrad8 | rad8∷hph | This study |
| SC196 Δubc13 | ubc13∷kanMX6 | Frampton et al (2006) |
| SC277 Δmms2 | mms2∷kanMX6 | Frampton et al (2006) |
| SC215 ΔdinB | dinB∷kanMX6 | This study |
| SC217 Δrev1 | rev1∷kanMX6 | This study |
| SC216 Δrev3 | rev3∷kanMX6 | This study |
| SC260 Δrad30 | rad30∷ kanMX6–nmt eso1 | This study |
| SC206 Δuve1 | uve1∷leu2 | Gift from P Russell |
| SC220 Δswi10 | swi10∷hph | This study |
| SC224 Δswi10 Δuve1 | swi10∷hph uve1∷leu2 | This study |
| SC229 Δswi10 Δuve1 Δmlh1 | swi10∷hph uve1∷leu2 mlh1∷kanMX6 | This study |
| SC256 Δswi10 Δuve1 Δmlh1 Δrhp18 | swi10∷hph uve1∷leu2 mlh1∷kanMX6 rhp18∷hph | This study |
| SC336 Δswi10 Δuve1 Δmlh1 pcn1-K164R | swi10∷hph uve1∷leu2 mlh1∷kanMX6 | This study |
| pcn1-K164R∷kanMX6 | ||
| SC241 Δswi10 Δuve1 Δmlh1 Δrad8 | swi10∷hph uve1∷leu2 mlh1∷kanMX6 rad8∷hph | This study |
| SC266 Δswi10 Δuve1 Δmlh1 Δubc13 | swi10∷hph uve1∷leu2 mlh1∷kanMX6 ubc13∷kanMX6 | This study |
| SC286 Δswi10 Δuve1 Δmlh1 Δmms2 | swi10∷hph uve1∷leu2 mlh1∷kanMX6 mms2∷kanMX6 | This study |
| SC272 Δswi10 Δuve1 Δmlh1 Δrad30 | swi10∷hph uve1∷leu2 mlh1∷kanMX6 rad30∷kanMX6 | This study |
| SC251 Δswi10 Δuve1 Δmlh1 ΔdinB | swi10∷hph uve1∷leu2 mlh1∷kanMX6 dinB∷kanMX6 | This study |
| SC252 Δswi10 Δuve1 Δmlh1 Δrev1 | swi10∷hph uve1∷leu2 mlh1∷kanMX6 rev1∷kanMX6 | This study |
| SC254 Δswi10 Δuve1 Δmlh1 Δrev3 | swi10∷hph uve1∷leu2 mlh1∷kanMX6 ubc13∷kanMX6 | This study |
| SC370 Δswi10 Δuve1 Δmlh1Δpol4 | swi10∷hph uve1∷leu2 mlh1∷kanMX6 pol4∷kanMX6 | This study |
| SC295 Δswi10 Δuve1 Δmlh1 Δrad30 ΔdinB | swi10∷hph uve1∷leu2 mlh1∷kanMX6 rad30∷ kanMX6 dinB∷kanMX6 | This study |
| SC359 Δswi10 Δuve1 Δmlh1 Δrad8 Δrad30 | swi10∷hph uve1∷leu2 mlh1∷kanMX6 rad8∷ kanMX6 rad30∷kanMX6 | This study |
| SC362 Δswi10 Δuve1 Δmlh1 Δrad8 Δrev1 | swi10∷hph uve1∷leu2 mlh1∷kanMX6 rad8∷ kanMX6 rev1∷kanMX6 | This study |
| SC363 Δswi10 Δuve1 Δmlh1 Δrad8 Δrev3 | swi10∷hph uve1∷leu2 mlh1∷kanMX6 rad8∷ kanMX6 rev3∷kanMX6 | This study |
| SC360 Δswi10 Δuve1 Δmlh1 Δrad30 Δrhp18 | swi10∷hph uve1∷leu2 mlh1∷kanMX6 rad30∷ kanMX6 rhp18∷hph | This study |
| SC378 Δswi10 Δuve1 Δmlh1 Δrad30 ΔdinB Δrev1 | swi10∷hph uve1∷leu2 mlh1∷kanMX6 rad30∷kanMX6 dinB∷kanMX6 rev1∷kanMX6 | This study |
| SC381 Δswi10 Δuve1 Δmlh1 Δrad30 ΔdinB Δrev3 | swi10∷hph uve1∷leu2 mlh1∷kanMX6 rad30∷kanMX6 dinB∷kanMX6 rev3∷kanMX6 | This study |
| SC383 Δswi10 Δuve1 Δmlh1 Δrad30 ΔdinB Δrev1 Δrev3 | swi10∷hph uve1∷leu2 mlh1∷kanMX6 rad30∷kanMX6 dinB∷kanMX6 rev1∷kanMX6 rev3∷kanMX6 | This study |
| All strains are derivatives of the ura-D18, leu1-32 genotype. | ||
We performed the colony-based TLS assay in strains that were deficient for the NER, UVR and MMR (num). In these strains, essential genes swi10, uve1 and mlh1 of the NER, UVR and MMR pathways were knocked out, respectively (Bahler et al, 1998; Sato et al, 2005). We confirmed the extreme sensitivity to low doses of UV of the num strain (Supplementary Figure S1A) and we verified that the PCNA ubiquitination profile of PRR mutants was not affected in a num background (Supplementary Figure S1B).
The MGZ E. coli strain (LacIq ΔlacZ(M15)lacY∷Tn10 mini-tet) is used for the TLS assay.
Plasmid construction
Single-adducted plasmids pSP-TT-CPD and pSP-TT(6-4) were constructed by ligation of an oligonucleotide containing either a single TT-CPD dimer or a TT-(6-4) photoproduct (5′-GCAAGTTAACACG) into a ‘gapped-duplex' plasmid constructed in vitro as described elsewhere (Burnouf et al, 1989; Becherel and Fuchs, 1999). These plasmids contain a short sequence heterology opposite the lesion site to allow independent assessment of the replication product of the two strands (Figure 1A). Covalently closed circular constructs are isolated by centrifugation on CsCl/ethidium bromide gradients. All plasmids are pUC derivatives containing a ColE1 origin and ampR cassette for E. coli manipulation. The plasmids also contain the bi-directional ARSDblet as replication origin in S. pombe cells (Brun et al, 1995) and the ura4 cassette to allow selection of transformants. The ARSDblet and Ura4 cassette were amplified by PCR and inserted at the unique NdeI site of the pCUL+ vector series. The UV lesion is located in the N-terminal region of the lacZ′ gene 300 nucleotides from the ARSDblet; thus, ensuring that the lesion is present in the leading strand during DNA replication.
TLS assay
S. pombe cell were grown in YES medium to OD600=0.5, then prepared for electroporation as described in Suga and Hatakeyama (2001). In all, 100 ng of single-lesion plasmid were electroporated into 200 μl freshly prepared competent cells (Figure 1B). Electroporated cells were then plated onto selective medium lacking uracil (LAH) to allow selection of cells that have received the plasmid. Plates were incubated at 30°C until colonies appeared (∼72 h). Transformed cells were collected and the total DNA extracted as described in Beach et al (1982) and subjected to DpnI digestion. Plasmid DNA was then transformed into MGZ E. coli strain and plated onto Lac indicator plates (X-Gal/IPTG/ampicillin/tetracyclin LB). After an incubation of 20 h at 37°C, the proportion of total TLS is determined as the number of blue colonies over the total number of colonies. Each determination is the average of three independent experiments resulting from the examination of several thousands colonies for each strain. The standard error of the mean (s.e.m.) is represented by the error bars.
To determine the percentage of mutagenic bypass, blue colonies are isolated and mini preparations of plasmid are performed and subjected to HpaI/XhoI double enzymatic digestion. Owing to the presence of the HpaI site (gttaac) at the lesion site and of the unique XhoI site, double enzymatic digestion generates a 1500 bp fragment if the replication of the single-lesion plasmid has occurred in a TLS error-free manner. In case of mutagenic bypass at the lesion site, the HpaI site is absent and the double enzymatic digestion generates a unique fragment of 5700 bp corresponding to the XhoI-linearized plasmid. The extent of error-free and mutagenic TLS is thus determined as a fraction of total TLS. The nature of the mutagenic TLS events is determined by sequence analysis using the following sequencing primer: 5′gcggtgtgaaataccgcacag.
Supplementary Material
Acknowledgments
We are very grateful to Joel Huberman and Takashi Toda for the gift of the pDblet and the pCR2.1 (−nat/hph) plasmids, respectively. We thank Paul Russell and Luis Blanco for the S. pombe strains. We thank Vincent Geli, Pierre-Henri Gaillard and Agnès Tissier for useful discussions. RF laboratory is supported by the ‘Agence Nationale de la Recherche (ANR-06-BLAN-0258). ARL laboratory is supported by the MRC and an EC RTN. SC is supported by the ‘Association pour la Recherche sur le Cancer' (ARC).
Footnotes
The authors declare that they have no conflict of interest.
References
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Beach D, Piper M, Nurse P (1982) Construction of a Schizosaccharomyces pombe gene bank in a yeast bacterial shuttle vector and its use to isolate genes by complementation. Mol Gen Genet 187: 326–329 [DOI] [PubMed] [Google Scholar]
- Becherel OJ, Fuchs RP (1999) SOS mutagenesis results from up-regulation of translesion synthesis. J Mol Biol 294: 299–306 [DOI] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I (2005) Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310: 1821–1824 [DOI] [PubMed] [Google Scholar]
- Bresson A, Fuchs RP (2002) Lesion bypass in yeast cells: Pol eta participates in a multi-DNA polymerase process. EMBO J 21: 3881–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun C, Dubey DD, Huberman JA (1995) pDblet, a stable autonomously replicating shuttle vector for Schizosaccharomyces pombe. Gene 164: 173–177 [DOI] [PubMed] [Google Scholar]
- Burnouf D, Koehl P, Fuchs RP (1989) Single adduct mutagenesis: strong effect of the position of a single acetylaminofluorene adduct within a mutation hot spot. Proc Natl Acad Sci USA 86: 4147–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton J, Irmisch A, Green CM, Neiss A, Trickey M, Ulrich HD, Furuya K, Watts FZ, Carr AM, Lehmann AR (2006) Postreplication repair and PCNA modification in Schizosaccharomyces pombe. Mol Biol Cell 17: 2976–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Lehmann AR, Fuchs RP (2005) Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell 18: 499–505 [DOI] [PubMed] [Google Scholar]
- Gangavarapu V, Haracska L, Unk I, Johnson RE, Prakash S, Prakash L (2006) Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Mol Cell Biol 26: 7783–7790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg P, Burgers PM (2005) Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1. Proc Natl Acad Sci USA 102: 18361–18366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs PE, McDonald J, Woodgate R, Lawrence CW (2005) The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics 169: 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Barrera S, Sanchez A, Ruiz JF, Juarez R, Picher AJ, Terrados G, Andrade P, Blanco L (2005) Characterization of SpPol4, a unique X-family DNA polymerase in Schizosaccharomyces pombe. Nucleic Acids Res 33: 4762–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC (2006) Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol 26: 8892–8900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A, Ziv O, Gueranger Q, Geacintov N, Livneh Z (2008) Reduced efficiency and increased mutagenicity of translesion DNA synthesis across a TT cyclobutane pyrimidine dimer, but not a TT 6-4 photoproduct, in human cells lacking DNA polymerase eta. DNA Repair (Amst) 7: 1636–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi K, Katayama T, Iwai S, Hidaka M, Horiuchi T, Maki H (2003) Fate of DNA replication fork encountering a single DNA lesion during oriC plasmid DNA replication in vitro. Genes Cells 8: 437–449 [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Haracska L, Prakash S, Prakash L (2001) Role of DNA polymerase zeta in the bypass of a (6-4) TT photoproduct. Mol Cell Biol 21: 3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Kondratick CM, Prakash S, Prakash L (1999) hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285: 263–265 [DOI] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR (2004) Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell 14: 491–500 [DOI] [PubMed] [Google Scholar]
- Lawrence CW, Gibbs PE, Borden A, Horsfall MJ, Kilbey BJ (1993) Mutagenesis induced by single UV photoproducts in E. coli and yeast. Mutat Res 299: 157–163 [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Kirk-Bell S, Arlett CF, Paterson MC, Lohman PH, de Weerd-Kastelein EA, Bootsma D (1975) Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc Natl Acad Sci USA 72: 219–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM (2006) Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 21: 15–27 [DOI] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Iwai S, Hanaoka F (2000) Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO J 19: 3100–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature 399: 700–704 [DOI] [PubMed] [Google Scholar]
- McInerney P, O'Donnell M (2004) Functional uncoupling of twin polymerases: mechanism of polymerase dissociation from a lagging-strand block. J Biol Chem 279: 21543–21551 [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S (2007) PCNA, the maestro of the replication fork. Cell 129: 665–679 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC (1996) Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science 272: 1646–1649 [DOI] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R (2001) The Y-family of DNA polymerases. Mol Cell 8: 7–8 [DOI] [PubMed] [Google Scholar]
- Pages V, Bresson A, Acharya N, Prakash S, Fuchs RP, Prakash L (2008) Requirement of Rad5 for DNA polymerase zeta-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics 180: 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages V, Fuchs RP (2003) Uncoupling of leading- and lagging-strand DNA replication during lesion bypass in vivo. Science 300: 1300–1303 [DOI] [PubMed] [Google Scholar]
- Parker JL, Ulrich HD (2009) Mechanistic analysis of PCNA poly-ubiquitylation by the ubiquitin protein ligases Rad18 and Rad5. EMBO J 28: 3657–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plosky BS, Vidal AE, Fernandez de Henestrosa AR, McLenigan MP, McDonald JP, Mead S, Woodgate R (2006) Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin. EMBO J 25: 2847–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L (2005) Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem 74: 317–353 [DOI] [PubMed] [Google Scholar]
- Sato M, Dhut S, Toda T (2005) New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast 22: 583–591 [DOI] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S (2009) Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J 28: 2461–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, Geacintov N, Livneh Z (2009) Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J 28: 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD (2003) Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191 [DOI] [PubMed] [Google Scholar]
- Suga M, Hatakeyama T (2001) High efficiency transformation of Schizosaccharomyces pombe pretreated with thiol compounds by electroporation. Yeast 18: 1015–1021 [DOI] [PubMed] [Google Scholar]
- Walters KJ, Chen X (2009) Measuring ubiquitin chain linkage: Rap80 uses a molecular ruler mechanism for ubiquitin linkage specificity. EMBO J 28: 2307–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M (2004) Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J 23: 3886–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M (2009) Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10: 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Prakash L, Prakash S (2009) Highly error-free role of DNA polymerase eta in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc Natl Acad Sci USA 106: 18219–18224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lawrence CW (2005) The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc Natl Acad Sci USA 102: 15954–15959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z, Johnson RE, Haracska L, Prakash L, Prakash S, Benkovic SJ (2008) Regulation of polymerase exchange between Poleta and Poldelta by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proc Natl Acad Sci USA 105: 5361–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv O, Geacintov N, Nakajima S, Yasui A, Livneh Z (2009) DNA polymerase zeta cooperates with polymerases kappa and iota in translesion DNA synthesis across pyrimidine photodimers in cells from XPV patients. Proc Natl Acad Sci USA 106: 11552–11557 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.