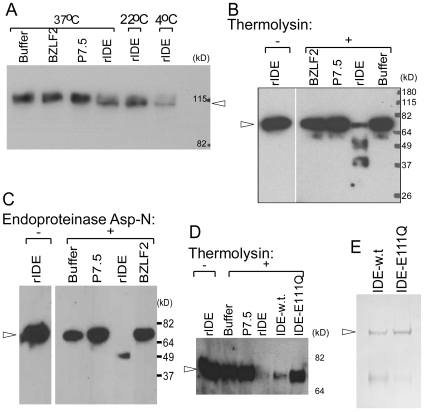
Figure 5. rIDE modifies gEt and induces a conformational change in gEt.
(A) Biotin labeled gEt-Fc protein (arrow) was incubated with rIDE at the indicated temperature for 30 min. After electrophoresis and transfer onto a nitrocellulose membrane, proteins were visualized with streptavidin-conjugated-horse radish peroxidase. (B) Biotin-labeled gEt-His protein (arrow) was incubated with buffer, control protein BZLF2 or P7.5, rIDE (produced in baculovirus) at 37°C for 30 min, followed by urea for 18 hr and thermolysin for 30 sec. The proteins were separated by electropheresis, transferred to a nitrocellulose membrane and proteins were detected using streptavidin-conjugated horse radish peroxidase. (C) Biotin-labeled gEt-His protein (arrow) was incubated with rIDE or control proteins at 37°C for 30 min, followed by incubation with 4 µg/ml of endoproteinase Asp-N (Roche Applied Science, Indianapolis, IN) at 37°C for 45 sec. The digestion was then terminated by adding 0.5 M EDTA and samples were boiled in SDS-PAGE gel loading buffer with 2.5% 2-mercaptoethanol and separated by electrophoresis. Protein fragments were detected by streptavidin conjugated-horse radish peroxidase. (D) Binding of catalytically inactive IDE mutant protein IDE-E111Q to gEt fails to induce a conformational change in gE. Biotin-labeled gEt-His protein (arrow) was incubated with rIDE (produced by baculovirus), IDE-E111Q (produced in bacteria), or IDE-w. t. (produced in bacteria), or negative control proteins as indicated at 37°C for 30 min, followed by thermolysin and processed as described in panel B. (E) Coomassie Blue stained SDS-PAGE gel showing the amount of IDE-E111Q and IDE-w.t. proteins used for pulse proteolysis assay in panel D.