Figure 2. NRDE-2 is recruited by NRDE-3/siRNA complexes to pre-mRNAs that have been targeted by RNAi.
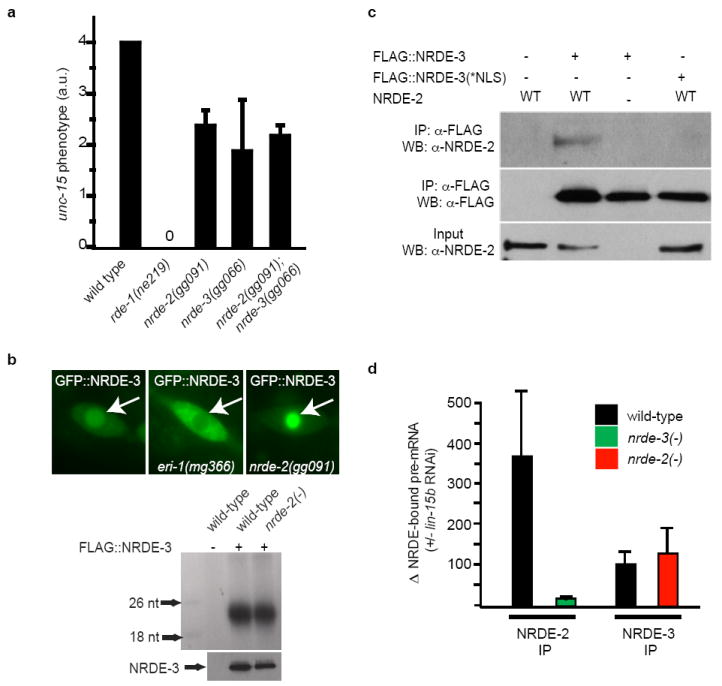
(a) Animals were exposed to unc-15 dsRNA and scored for uncoordinated phenotypes (Unc) (n=3, +/- s.d.). rde-1(ne219) animals are defective for RNAi 12. (b) (top panels) Fluorescent microscopy of a seam cell expressing GFP∷NRDE-3. Arrows indicate nuclei. eri-1(mg366) animals fail to express endo siRNAs and consequently NRDE-3 is mislocalized to the cytoplasm 3,13,14. (bottom panels) FLAG∷NRDE-3 co-precipitating RNAs 32P-radiolabeled and analyzed by PAGE. (c) NRDE-2 co-precipitates with nuclear-localized NRDE-3 (materials and methods) (n=3). (d) qRT-PCR quantification of NRDE-2/3 co-precipitating pre-mRNA. Throughout this manuscript pre-mRNA levels are quantified with exon/intron or intron/intron primer pairs. Data are expressed as ratios of co-precipitating pre-mRNA +/- lin-15b RNAi (n=4 for NRDE-2 IP, n=2 for NRDE-3 IP, +/- s.e.m.). Δ=fold change.
