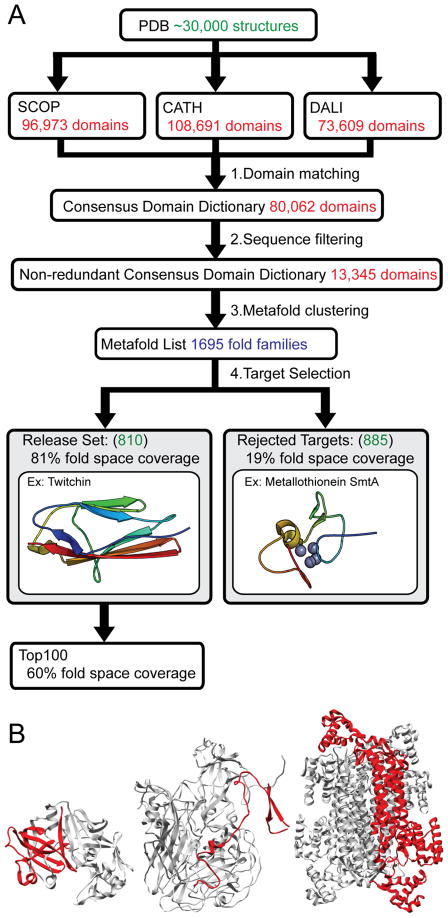
Figure 1.
Consensus domain dictionary generation and target selection. (A) Domain dictionaries partition structures from the Protein Data Bank into domains and folds. The current versions of the SCOP, CATH and DALI domain dictionaries include about 30,000 structures. We find consensus between the domains within these separate domain dictionaries to generate consensus domains. These consensus domains are filtered by sequence to generate a non-redundant consensus domain dictionary. These non-redundant domains are then clustered into metafolds and ranked by population. A single fold representative is selected from each metafold. This representative is either suitable for simulation and becomes part of our release set, or is judged unsuitable for simulation and is rejected. Simulation and analysis data of fold representatives from the Top 100 most populated folds in our release set are publicly available on our website. (B) Three examples of fold representatives (in red) that were rejected because they are not truly autonomous domains. Left, cathepsin D from PDB 1LYA; Middle, chain 4 in the human poliovirus 1, from PDB 1AL2; Right, delta crystallin I from PDB 1I0A.