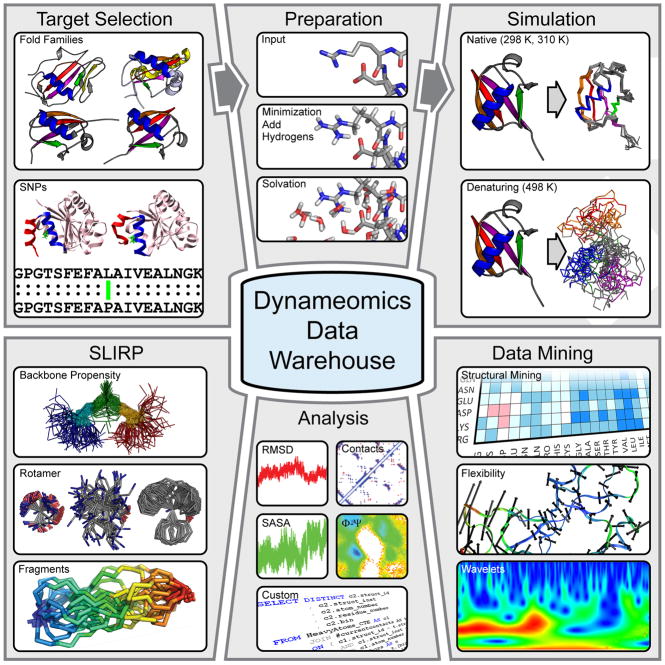
Figure 2.
Overview of the generation and use of our simulation data, organized through the Dynameomics database. After selection of protein targets to cover the majority of known protein folds (Figure 1) as well as to examine the influence of single residue mutations (Figure 5), structures obtained from the PDB are prepared for simulation by addition of hydrogens, minimization and solvation, according to our standard protocols (see text). Simulations are then run to capture both native state and unfolding dynamics. The simulation trajectories are stored in the database, together with a set of standard analysis (which can be viewed online). The simulations of native state dynamics are used to define global residue properties and build a fragment library (Figure 3). Specialized mining of the simulations in the database is also performed in-house, to examine further questions on protein dynamics and unfolding (Figure 4), including the use of new techniques such as wavelet and flexibility analysis. We also offer direct access to the native state simulations of the Top 100 fold representatives in our database to external users, who can query these data using SQL Server.