Figure 6.
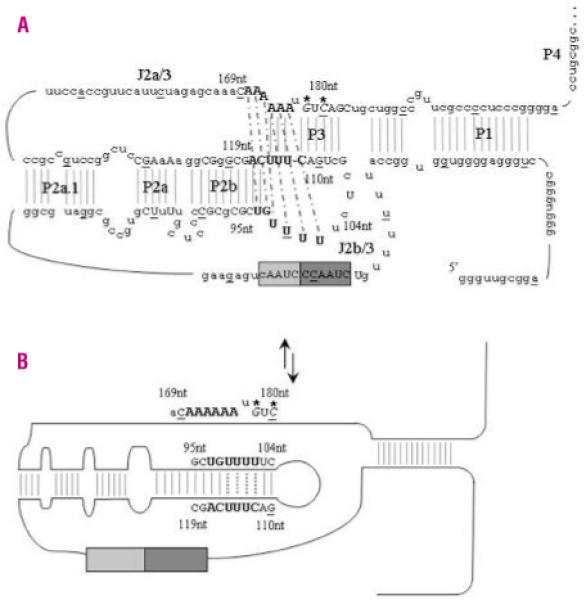
TERC pseudoknot structure and proposed molecular switch model. A. The secondary structure of the TERC pseudoknot has been elucidated by functional analysis. Every tenth base of the 451bp molecule is underlined and <80% vertebrate nucleotide conservation is denoted by upper case letters. The RNA template contains partial repeats of the telomere repeat known as the alignment region (lighter gray box). The larger dark grey box contains the templating region which is utilized to ensure correct base-pairing and correct translocation and re-alignment for the next round of template synthesis. Critical bases involved in the triple helix structure are highlighted in bold. Thermodynamics and mutational analysis suggest that nucleotides 171-172AA form a Hoogsteen base-pair with 97-98UG, which are helically-bound by conventional Watson-Crick base-pairing to 116-117CA. Nucleotides 100-102UUU then form a Hoogsteen base-pair with 174-176AAA, which are helically-bound by conventional Watson-Crick base-pairing to 113-115UUU. Nucleotides 99U and 173A can then pair up between these two triple helices to bridge the physical gap. The Watson-Crick base-pairing is denoted by gray lines while the dashed black lines represent the Hoogsteen basepairing. The de novo mutations described in this paper are in italics and are highlighted with *. B. While the two novel de novo mutations are not directly involved in the loop formed during the proposed molecular switch, the complementary nucleotides G110 and C112 are critical to the correct formation of the new TERC RNA structure.
