Figure 2. A map of viral-human protein interactions identifies a dense interconnected network, coupled to key cellular signaling pathways. (A) Viral-viral interactions.
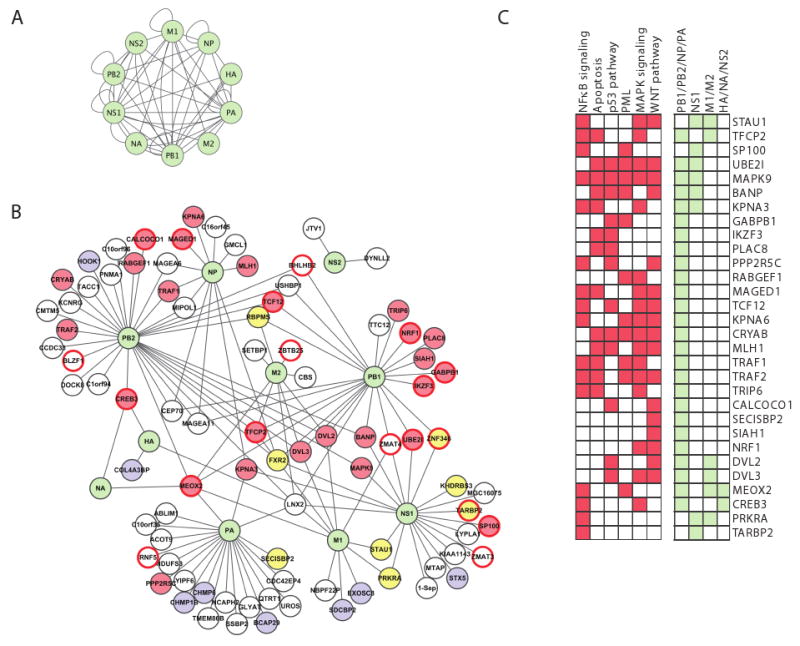
Green nodes, viral proteins; edges, direct physical interactions observed in a Y2H assay. (B) Influenza-human interactions. 134 interactions (edges) connect the ten influenza proteins (green nodes) to 87 ‘H1’ human proteins. Yellow fill, RNA-binding proteins; blue fill, protein transport; red border, transcription factors; red fill, 30 proteins that play a role in four major signaling pathways (NFκB, apoptosis, MAPK and WNT signaling); white fill, proteins with other functions. (C) 30 host interactor proteins (H1) are shown with their membership in specific pathways (red) or direct interactions with influenza proteins (light green). H1 proteins are either known components or established direct interactors with components of these pathways (see Experimental Procedures). Many of the 30 proteins are involved in multiple signaling pathways, and interact with polymerase subunits. Description of influenza A proteins: PB1, PB2, PA form the viral polymerase; NP complexes with viral RNA and is required for viral polymerase activity on RNA; HA and NA are membrane proteins involved in the fusion and release of viral particles; M1 is a matrix protein involved in export and assembly of RNA and viral particles; M2 modulates fusion through its ion channel activity; NS1 is a non-structural protein that regulates host pathways; NS2 is involved in RNA export.
