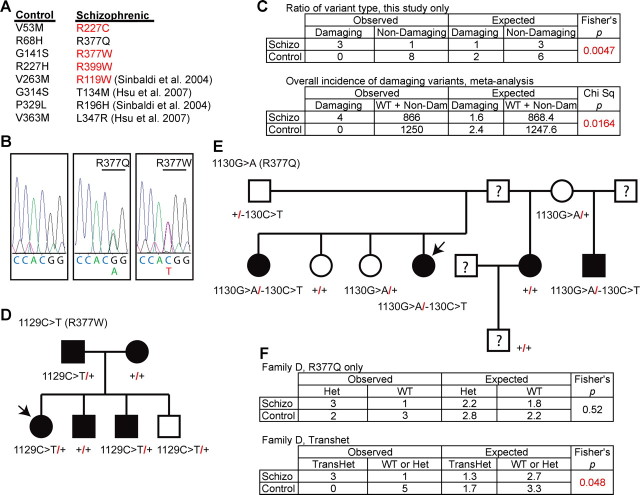
Figure 2.
NGR locus and genetic variation in schizophrenia. A, The NGR coding sequence from 542 schizophrenic individuals and 650 healthy controls was determined. The nonsynonymous coding sequence variants listed were present heterozygously in 12 individuals. Those variants listed in red are predicted to be “probably damaging” by the PolyPhen program. In addition, the four nonsynonymous coding sequence variants from two previous studies containing 328 schizophrenic individuals and 600 healthy controls are cited. B, 5′–3′ DNA sequence traces from a control individual with two WT NGR alleles, an affected individual heterozygous for a G>A transition at nucleotide 1130 resulting in the R377Q substitution, or an affected individual heterozygous for a C>T transition at nucleotide 1129 resulting in the R377W substitution. C, The observed numbers of coding variants predicted to be “probably damaging” or not by PolyPhen (http://genetics.bwh.harvard.edu/pph/) (Sunyaev et al., 2001) in the two groups is tabulated and analyzed statistically for the existing study. The metaanalysis table includes the individuals from previous studies and assesses the overall incidence of “probably damaging” variants in the two populations. D, E, Pedigree for R377W and R377Q families. Schizophrenic probands are indicated by the arrows. The filled symbols represent individuals with major psychiatric disease: schizophrenia, major depression with psychotic features, mild mental retardation with schizoid personality, or manic-depressive illness. F, The occurrence of R377Q variant or the transheterozygous R377Q variant in the presence of the 5′ noncoding change is tabulated for the family in D.