Abstract
The nitrogenase Fe protein contains a [4Fe-4S] cluster and delivers one electron at a time to the catalytic MoFe protein. During this electron delivery, the Fe protein in its [4Fe-4S]1+ reduced state (Fered) binds two MgATP and forms a complex with the MoFe protein, with subsequent transfer of one electron to the MoFe protein in a reaction coupled to the hydrolysis of two ATP. Crystal structures with the nitrogenase complex in different nucleotide-bound states show major conformational changes which provide a structural underpinning to suggestions that inter-component electron transfer (ET) is ‘gated’ by conformational changes of the complex and/or of its component proteins. Although electron delivery is coupled to ATP hydrolysis, their connection is puzzling, for it appears that ET precedes both ATP hydrolysis and Pi release. We here test the gating hypothesis with studies of the intracomplex oxidation of Fered by MoFe protein in the presence of a variety of solutes. Conformational control of this process (gating) is revealed by the finding that it responds to changes in osmotic pressure (but not viscosity), with no fewer than 80 waters being bound during the reaction. The absence of a solvent kinetic isotope effect further implies that ATP hydrolysis does not occur during the rate-limiting step of ET.
The Mo-dependent nitrogenase1,2 is comprised of two component proteins, denoted the Fe and MoFe protein. The Fe protein contains a [4Fe-4S] cluster and delivers one electron at a time to the MoFe protein, which contains the multimetallic FeMoco catalytic cluster [7Fe, Mo, 9S; X], as well as an auxiliary [8Fe, 7S] P cluster that might be mediating electron transfer from Fe protein to FeMo-co. During electron delivery, the Fe protein in its [4Fe-4S]1+ reduced state (Fered) binds two MgATP and rapidly forms a complex with the MoFe protein; subsequent transfer of one electron to the MoFe protein is coupled to ATP hydrolysis.
Crystal structures with the nitrogenase complex in different nucleotide-bound states show that major conformational changes occur upon ATP hydrolysis, Fig 1.3,4 This provides a structural underpinning to suggestions1,5,6 that inter-component electron transfer (ET) is ‘gated’7-9 by conformational changes of the complex and/or of its component proteins. Although electron delivery is coupled to ATP hydrolysis, their connection is puzzling, for it appears that ET precedes both ATP hydrolysis10 and Pi release.11 Furthermore, none of the X-ray structures of the Fe protein-MoFe protein complex, [Fe: MoFe], reveals any perturbation of either of the two clusters within the MoFe protein that might produce conformational activation of electron transfer.4
Figure 1.
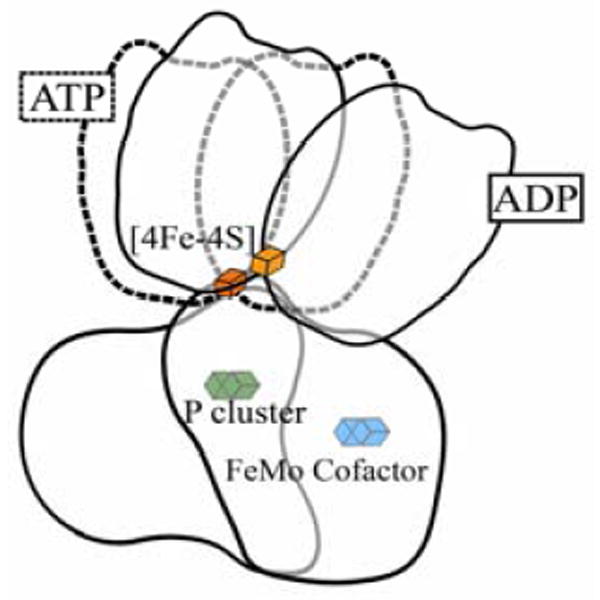
Representation of complexes between Fe protein (top) and MoFe protein (bottom) in ATP- like (dashed) and ADP-like bound forms; (adapted from Tezcan and coworkers3).
To determine whether conformational changes control ET within the [Fe: MoFe] protein complex, we have measured oxidation of Fered by MoFe protein in the presence of solutes that increase the viscosity (η) and osmotic pressure of the solution. The rate constant for a dynamical conformational transition varies with viscosity as, k(η) ∝ 1/η.12 Changes in osmotic pressure instead modulate the energetics of reactions that change the number of bound waters.13 The rate constant for such a process varies exponentially with the molality (m) of added solute according to the equation,13 k(m) ∝ exp[-(Δn/55.6)m], where Δn is the number of waters absorbed in the transformation.
Figure 2 shows typical traces that monitor the oxidation of Fered by resting-state MoFe protein in solutions with varying concentrations of sucrose as viscogen/osmolyte. The ET reaction was initiated by the addition of MgATP to a solution of [Fered] and [MoFe] in the stopped flow. MgATP was added in sufficient concentrations that the known association constants17 ensure that Fered binds two ATP and binds to MoFe within the dead-time of the instrument.14 As a result, the absorbance changes in these pre-steady-state experiments are wholly associated with Fered oxidation within the [Fered(MgATP)2: MoFe] complex.15,16
Figure 2.
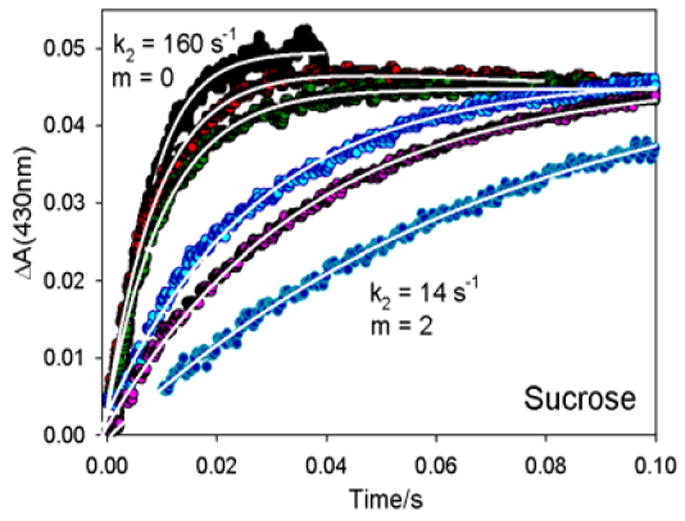
Stopped-flow oxidation of Fered within [Fered(MgATP)2: MoFe] complex. Sucrose concentrations vary from 0 to 2 m. Traces for m = 0 in H2O and D2O overlay so that the latter would barely be visible if shown, and hence is not (traces have same k2 within error). Conditions: 75 μM Fe protein, 20 μM MoFe protein; 100 mM HEPES buffer, pH 7.4: 25 C.
The stopped-flow absorbance increases, caused by the oxidation of Fered, are exponential (Fig 2); k2 = 160(10) s-1 for aqueous buffer is in excellent agreement with previous measurements.1,17 Progressive additions of either sucrose (Fig 2), glucose, raffinose, PEGs 300, 600, or glycerol all cause progressive decreases in k2. This solute control of ET reveals that intra-complex oxidation of Fered by resting-state MoFe protein is indeed ‘gated’ by a conformational transition that activates ET.
Figure 3 presents logarithmic plots of k2 vs molality for each solute employed. Each plot is linear, revealing that osmotic pressure effects generate the changes in k2. This is confirmed by noting that, in all cases, the changes are larger than could be generated by viscosity effects alone. For example, with sucrose as solute, for m = 2 the increased viscosity could at most decrease k2 by 1/ η ≈ 1/5, whereas k2 decreases by over 1/10; more dramatically, for glycerol as solute, with m = 3.2 the increased viscosity could at most decrease k2 by ~1/2, whereas k2 decreases by ~ 1/15. In fact, it appears that viscosity plays no role in the solute-induced changes in ET. The plots of k2 vs m for PEG300 and 600 completely overlay, but the viscosities differ by roughly a factor of two at any given molality.
Figure 3.
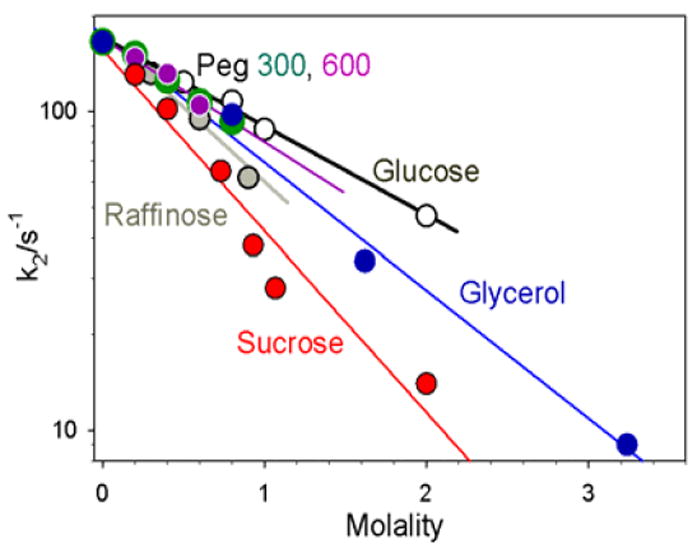
Log of the rate constants for oxidation of Fered within the [Fered(MgATP)2: MoFe] complex as a function of osmolyte concentration.
When k2 is controlled by osmotic pressure effects, the slope of the plot of log k vs m is proportional to [-Δn] for the conformational conversion (see above).13 The negative slopes in Fig 3 thus correspond to an uptake of waters, Δn > 0. Multiple osmolytes were employed because the value of Δn measured in osmotic pressure experiments in general differs among solutes because they can interact with the protein surface in different ways, displacing different numbers of bound waters and yielding different apparent values for Δn.13 The osmolyte whose slope is greatest interacts least and displaces the fewest waters, thereby giving the best value for Δn. In this study that osmolyte is sucrose,18 and its slope in Fig 3 gives, Δn ~ + 80 as the best available lower-bound value for the number of waters that bind to newly exposed surfaces during the conformational transition.
Taking roughly one water to be bound per ΔA0 ~ 10 Å2 of exposed surface, the binding of Δn ~ 80 waters would correspond to a conformational transition in which ΔA ~ 800 Å2 of surface becomes exposed. To calibrate this value, the ‘ADP’ structure of the [Fe: MoFe] complex (Fig 1) exposes ~ 2000 Å2 more protein surface than does the ‘ATP’ structure.3,4 Thus, the conformational change revealed by the present measurements can be plausibly attributed to a large-scale motion of the Fe protein relative to MoFe protein, such as in Fig 1, but likely one that is of somewhat lesser extent and thus exposes correspondingly less surface. However, one cannot rule out other types of structural changes.
ATP hydrolysis is accompanied by the release of 0.5 protons/ATP,10 so we measured the solvent kinetic isotope effect, sKIE = k2(H2O)/k2(D2O), for the gated intracomplex Fered oxidation to test if the rate-limiting step involves proton transfer. If this were the case one would expect sKIE > 1, as seen for ATP hydrolysis by the F1 ATPase.19 Instead, we find that the oxidation of Fered has no kinetic isotope effect: sKIE = 1, within error (see caption, Fig 2), an indication that ATP hydrolysis is not involved in the rate-limiting step of Fered oxidation.
How does conformational activation facilitate ET? One possibility is that it generates a transition state for Fered oxidation whose structure is optimized for direct ET from the [4Fe-4S]1+ cluster of Fered to FeMo-co. If the transition state occurs along an (imagined) reaction coordinate whose beginning and end points are the ATP-like and ADP-bound structures, Fig 1, it is unlikely that the conformational changes would enhance ET by decreasing the donor-acceptor distance: this transition increases the distances from the [4Fe-4S] cluster of the Fe protein to both the P cluster and FeMo-co.3 Among other possibilities, one may imagine that the Δn waters being bound include ordered water in the interface of the active complex, and that those enhance ET.20
Alternatively, one would expect an absence of viscosity effects if the solute effects were indeed wholly energetic, and did not operate on a dynamical process. This could occur if the rate-limiting step were preceded by a rapid pre-equilibrium between the energetically favored structure of the ATP-bound form of the complex and a higher-energy structure activated for ET, eq 1:
| (1) |
The gating limit embodied in eq 17 has been termed ‘conformational coupling’.9 If the activated structure binds Δn additional waters, then K*, and thus the observed ET rate constant, k2 = kK*, would be independent of viscosity but vary exponentially with the osmolyte molality (m), as is seen (Fig 3).13
Experiments are under way to test the assignment of the gating motions to rearrangements such as those in Fig 1, the alternate mechanisms noted above, and the role of the P cluster. It seems likely to us that the nitrogenase complex employs a ’compound’ ET gate, with gating motions revealed here being accompanied by as yet unknown conformational changes, at least some within the MoFe protein, which trigger events such as intra-MoFe ET, ATP hydrolysis,10 Pi release,11 and dissociation of the complex.
Acknowledgments
This work has been supported by the NIH (GM59087, LCS and DRD; HL63203, BMH). The authors thank Zhao Li for technical assistance.
References
- 1.Burgess BK, Lowe DJ. Chemical Reviews (Washington, D C) 1996;96:2983–3011. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 2.Seefeldt LC, Hoffman BM, Dean DR. Annu Rev Biochem. 2009;78:701–722. doi: 10.1146/annurev.biochem.78.070907.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tezcan FA, Kaiser JT, Mustafi D, Walton MY, Howard JB, Rees DC. Science (Washington, DC, U S) 2005;309:1377–1380. doi: 10.1126/science.1115653. [DOI] [PubMed] [Google Scholar]
- 4.Rees DC, Tezcan FA, Haynes CA, Walton MY, Andrade S, Einsle O, Howard JB. Phil Trans R Soc A. 2005;363:971–984. doi: 10.1098/rsta.2004.1539. [DOI] [PubMed] [Google Scholar]
- 5.Lanzilotta WN, Parker VD, Seefeldt LC. Biochemistry. 1998;37:399–407. doi: 10.1021/bi971681m. [DOI] [PubMed] [Google Scholar]
- 6.Syrtsova LA, Kotel’nikov AI, Shkondina NI, Lapshina MA. Kinet Catal. 2004;45:31–39. [Google Scholar]
- 7.Hoffman BM, Ratner MR. J Am Chem Soc. 1987;109:6237–6243. [Google Scholar]
- 8.Patel AD, Nocek JM, Hoffman BM. J Phys Chem. 2008;112:11827–11837. doi: 10.1021/jp8054679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson VL. Acc Chem Res. 2008;41:730–738. doi: 10.1021/ar700252c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorneley RNF, Ashby G, Howarth JV, Millar NC, Gutfreund H. Biochem J. 1989;264:657–661. doi: 10.1042/bj2640657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowe DJ, Ashby GA, Brune M, Knights H, Webb MR, Thorneley RNF. In: Nitrogen Fixation: Fundamentals and Applications. Tikhonovich IA, editor. Kluwer Academic Publishers; 1995. pp. 103–108. [Google Scholar]
- 12.Steinfeld JI, Francisco JS, Hase WL. Chemical Kinetics and Dynamics. 2. Prentice-Hall, Inc.; Upper Saddle River, New Jersey: 1999. [Google Scholar]
- 13.Parsegian VA, Rand RP, Rau DC. Methods Enzymol. 1995;259:43–94. doi: 10.1016/0076-6879(95)59039-0. [DOI] [PubMed] [Google Scholar]
- 14.Oxidation of Fered to Feox was followed at 430 nm with a Hi-Tech Instruments stopped-flow system placed inside an Ar-filled glove box. To compensate for absorbance changes from MgATP binding to Fered, a trace collected by mixing Fered with the MgATP solution was subtracted from each experimental trace. Fe and MoFe proteins were purified as described: Christiansen J, Goodwin PJ, Lanzilotta WN, Seefeldt LC, Dean DR. Biochemistry. 1998;37:12611–12623. doi: 10.1021/bi981165b.
- 15.Lowe DJ, Fisher K, Thorneley RNF. Biochem J. 1993;292:93–98. doi: 10.1042/bj2920093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher K, Lowe DJ, Tavares P, Pereira AS, Huynh BH, Edmondson D, Newton WE. J Inorg Biochem. 2007;101:1649–1656. doi: 10.1016/j.jinorgbio.2007.07.037. [DOI] [PubMed] [Google Scholar]
- 17.Wilson PE, Nyborg AC, Watt GD. Biophys Chem. 2001;91:281–304. doi: 10.1016/s0301-4622(01)00182-x. [DOI] [PubMed] [Google Scholar]
- 18.Indeed, sucrose behaves as an ideal solute for osmotic pressure studies, as its H-bonds with water have similar energetics to water-water H-bonds and it does not change the apparent density of water: Dougherty RC. J Phys Chem B. 2001;105:4514–4519.
- 19.Urbauer JL, Dorgan LJ, Schuster SM. Arch Biochem Biophys. 1984;231:498–502. doi: 10.1016/0003-9861(84)90413-2. [DOI] [PubMed] [Google Scholar]
- 20.Lin J, Balabin IA, Beratan DN. Science (Washington, DC, U S) 2005;310:1311–1313. doi: 10.1126/science.1118316. [DOI] [PMC free article] [PubMed] [Google Scholar]


