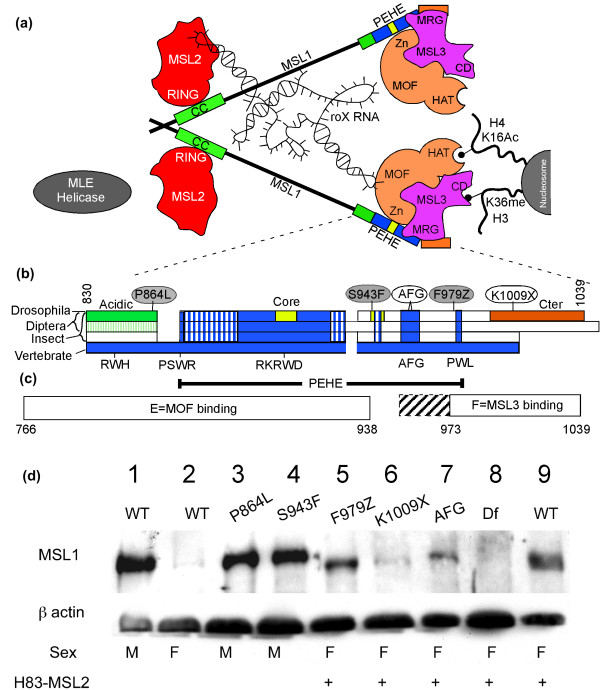
Figure 2.
Male Specific Lethal (MSL)1 modifier mutants map to the conserved C-terminal PEHE domain. (a) MSL subunit interactions found in previous biochemical and genetic analysis [5,23,25,51,55-57]. CC = coiled coil; HAT = histone acetyltransferase; Zn = Zn finger. Much of the MSL1 sequence is poorly conserved between Drosophila species (thin line). MSL1 is portrayed forming dimers at its N-terminus [24]. The chromodomain (CD) of MSL3 has been postulated to bind RNA [58], but here is depicted as binding histone H3 K36me [52,53]. Interactions between RNA and MSL proteins are reported in [26-29,31,58]. (b) The sequence alignment presented in Additional file 2 is illustrated graphically. First bar = 12 Drosophila. Second bar = three mosquitoes. Third bar = five non-dipteran insects. Fourth bar = 10 vertebrates. When other species share similarity to vertebrates (blue) they are also coloured blue or hatched blue for weak similarity. Open boxes = dissimilar sequences. Three regions are strongly conserved within Drosophila, but not found in any other group: a highly acidic region before the PEHE (green), PEHE core domain (yellow), and Cter (orange). Locations of the three modifier alleles (red) and two non-modifier alleles (white) are shown above. The lower labels show the four tryptophan residues and the AFG triplet that serve as sequence landmarks. The PEHE domain (Marin [50]) is shown below. (c) Morales et al. [25] showed MSL1 fragment E was sufficient to bind MOF ('males absent on first') and fragment F could bind MSL3. We found additional MSL3 contacts occur upstream of codon 979 including the AFG triplet (hatched box). (d) Anti-MSL1 western blot. Lane 1, wild-type (wt) male; 2, wt female; 3, msl1P864L male; 4, msl1S943F male; lanes 5-9, females expressing ectopic MSL2. Females make less MSL1 protein than males [46]; 5, msl1F979Z; 6, msl1K1009X; 7, msl1AFG; 8, msl1L60; 9, msl1L60/CyO. Loading control, β-actin.