Table 3.
Me-lex induced Hprt mutation spectrum in EM-C11
| Mutant/(s) | cDNA producta | Position | Type of mutation | Target | ||
|---|---|---|---|---|---|---|
| sequenceb | Aminoacid change N° of | |||||
| (exon) | (5’ > 3’) | (or | ||||
| cDNA change) | mutants | |||||
| Base pair changes | ||||||
| EN-15A | full length | 110 (2) | TA>CG | GTG TTT | ||
| ATT CCT CAT | Ile>Thr | 1 | ||||
| EN-11A | full length | 296 (3) | TA>GC | GTA GAT | ||
| TTT ATC AGA | Phe>Cys | 1 | ||||
| EN-20C | full length | 437 (6) | TA>GC | CAA ACT | ||
| CTG CTT TCC | Leu>Arg | 1 | ||||
| EN-25C | full length | 466 (6) | AT>TA | AAC CCC | ||
| AAA ATG GTT | Lys>STOP | 1 | ||||
| EN-5A | full length | 479 (6) | TA>AT | GTT AAG | ||
| GTT GCA AGC | Val>Asp | 1 | ||||
| EN-6B | full length | 543 (8) | TA>GC | GTT GGA | ||
| TTT GAA ATT | Phe>Leu | 1 | ||||
| EG-18B | full length | 545 (8) | AT>GC | GGA TTT GAA | ||
| ATT CCA | Glu>Gly | 1 | ||||
| Frameshift mutations | ||||||
| EN-29A | low, full length | 294–297 (3) | −T | CT GTA GAT | ||
| TTT ATC AG | 1 | |||||
| EN-19B | low, full length | 408–409 (6) | +AA | AG GAC ATA | ||
| ATT GAC AC | 1 | |||||
| Splice mutations | ||||||
| EN-28A | short cDNA | In 3: +1 | GC>AT | TACTGT g | ||
| taagtataatt | Skipping ex 3 | 1 | ||||
| EN-2B/−14C | short cDNA | In 3: −1 | GC>CG | ttttttaacta g | ||
| AATGAT | Skipping ex 4 | 5 | ||||
| /−17B/−21A | ||||||
| /−27C | ||||||
| EN-30B | short cDNA | In6: +2 | TA>GC | TGCAAGg t | ||
| atgtatgcc | Skipping ex 6 | 1 | ||||
| EN-3C/−7B | short cDNA | Splice junctions not sequenced | ||||
| Skipping ex 2 | 2 | |||||
| All exons present in gDNA | ||||||
| Genomic deletions | ||||||
| EN-1C | full length | 81–83 (2) | -CTA | AAT CAC | ||
| TAT GTC GAG | -Tyr | 1 | ||||
| EG-3A/−30A | full length | 565–567 (6) | -GTT | TTT GTT GTT | ||
| GGA TAT | -Val | 2 | ||||
| EN-10A | low, two short cDNAs | Deletion of exon 3 | ||||
| 1 | ||||||
| EG-13B | no cDNAs | Deletion of exon 4 | ||||
| 1 | ||||||
| EG-21B | no cDNAs | Deletion of exon 5 | ||||
| 1 | ||||||
| EG-2B/−8B | no cDNAs | Deletion of all Hprt exons | ||||
| 5 | ||||||
| /−24C/−27B | ||||||
| /−28C | ||||||
| Mutations in regulatory regions | ||||||
| EN-12A/−24A | no cDNAs | All exons present on gDNA | ||||
| 2 | ||||||
| EN-22B | very low, short cDNA | All exons present on gDNA | ||||
| Skipping ex 4 | 1 | |||||
| No mutations found at splice junctions | ||||||
| EG-4A/−5C | low ex (1–4) | All exons present on gDNA | ||||
| 8 | ||||||
| /−16A/−20B | ||||||
| /−21A/−22C | ||||||
| /−25A/−26C | ||||||
| EG-6B | Low gDNA ex 1. All other exons present on | |||||
| gDNA | 1 | |||||
| EG-9A | low cDNA ex (3–9) | All exons present on gDNA | ||||
| 1 | ||||||
| EG-12A/−19B | no cDNAs | All exons present on gDNA | ||||
| 3 | ||||||
| /−29A | ||||||
| Other mutations | ||||||
| EN-23A | two cDNA: full length, | Deletion (28–32) in exon 2 | tttttcag | |||
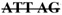 T GAT T GAT |
1 | |||||
| short | ||||||
| Skipping ex 2 | ||||||
cDNA amplifications of the full length, the 5'- (ex 1–4) and the 3'- (ex 4–8) half of the Hprt coding sequence were performed for each mutant (see M&M). Short cDNA refers to PCR fragments lacking the internal part of the hprt coding sequence.
Non transcribed strand gDNA = genomic DNA
