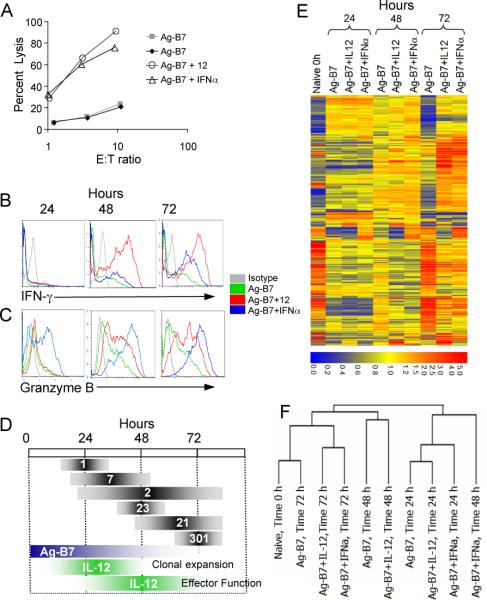
Figure 1. Programming of CD8 T cell differentiation requires IL-12 and/or IFNα along with Ag and costimulation.
(A) Purified naïve (CD44lo) OT-1 CD8 T cells were stimulated for 3 days in vitro with aAPC coated with H-2Kb / Ova257-264 peptide and B7-1 either alone, or with IL-12 or IFNα. All cultures were supplemented with IL-2. Cytolytic activity was measured on day 3 by 51Cr release assay using E.G7 target cells.
(B-C) Cells stimulated as in (A) were analyzed by intracellular staining on days 1, 2 and 3 for expression of IFNγ and grzB.
(D) The upper bars (gray) show the numbers of genes regulated in common by IL-12 and IFNα at 24, 48 and 72h. The lower bars (colored) show the times during which Ag-B7 and IL-12 must be present for optimal clonal expansion and development of effector function (from ref. (16)).
(E) Expression patterns for 408 probe ids representing the 375 genes regulated by both IL-12 and IFNα. The stimuli used and time in culture for each sample are shown at the top. Red represents high expression (signal value) and blue represents low expression on a log2 scale of 6.0 to 0.0 as shown at the bottom. Data not meeting the selection criterion was excluded and appears gray. The signal value was normalized to around 1.
(F) Hierarchical clustering linkage for cells treated with the various stimuli at 0, 24, 48 and 72h.