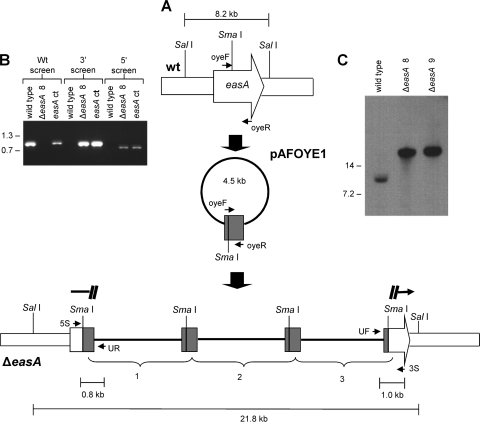
FIG. 2.
easA disruption strategy. (A) The disruption construct pAFOYE1 contains an incomplete, internal fragment of easA coding sequences (amplified from primers oyeF and oyeR) and was linearized at its sole SmaI site prior to transformation. The lower portion of this diagram shows tandem integration of three constructs in an easA disruptant (ΔeasA). The primer annealing sites are as follows: UF, universal (forward) primer sequences in the vector; 3S, primer flanking the 3′ border of the integration site; UR, primer derived from reverse primer sequences in the vector; 5S, primer flanking the 5′ border of the integration site. wt, wild type. (B) Primers 5S and 3S primed amplification of a 1.0-kb fragment only in the wild-type isolate. Primers UF and 3S primed amplification of the indicated 1.0-kb product from ΔeasA disruptant 8 (ΔeasA 8) and an easA complemented strain (easA ct). Primers UR and 5S primed amplification of the expected 0.8-kb band from strains with a disrupted easA allele (ΔeasA disruptant 8) and the ectopically complemented strain but not from the wild type. Relative mobilities of relevant fragments (sizes in kb) of BstEII-digested bacteriophage lambda are indicated. (C) Integration of transforming DNA at only the targeted site, as confirmed by Southern blot hybridization. DNA was digested with SalI and hybridized with a digoxigenin-dUTP-labeled easA probe. Relative mobilities of relevant fragments (sizes in kb) of BstEII-digested bacteriophage lambda are indicated. ΔeasA 9, ΔeasA disruptant 9.