Fig. 1.
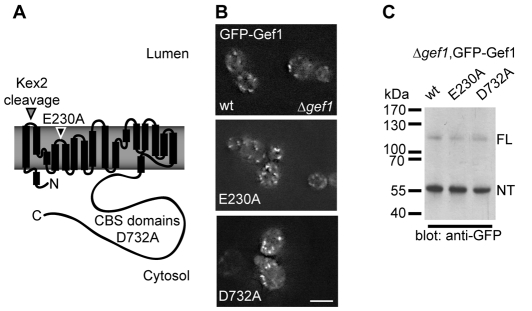
Structure-based mutagenesis of GEF1. (A) Scheme of the Gef1 protein based on helices present in the crystal structure of the prokaryotic homologue from E. coli (Dutzler et al., 2002). A Kex2 protease-cleavage site in the first extracellular loop is indicated. Kex2 is known to process Gef1 in the late Golgi, yielding a smaller N-terminal and a large C-terminal fragment. The position of the gating glutamate E230 is indicated. In closely related mammalian homologues such as ClC-5, mutation of the gating glutamate to alanine leads to uncoupled chloride movement through the transporter (Picollo and Pusch, 2005; Scheel et al., 2005). The C-terminus of eukaryotic CLC homologues contains cystathione β-synthetase (CBS) domains that have been shown to bind ATP in a recent crystal structure of the C-terminus of ClC-5 (Meyer et al., 2007). In ClC-5, mutation of the aspartate corresponding to Gef1 D732 to alanine abolishes ATP binding. (B) Subcellular localisation of N-terminal GFP-Gef1 fusions with the indicated mutations revealed by live-cell imaging in a Δgef1 strain. Scale bar: 5 μm. (C) Anti-GFP western blot of total cellular lysates from cells expressing the indicated GFP-Gef1 fusion proteins. ‘FL’ and ‘NT’ respectively indicate the migration position of uncleaved full-length Gef1 and the N-terminal product of the cleavage reaction.