Abstract
Herein we firstly reported a simple, environment-friendly, controllable synthetic method of CuSe nanosnakes at room temperature using copper salts and sodium selenosulfate as the reactants, and bovine serum albumin (BSA) as foaming agent. As the amounts of selenide ions (Se2−) released from Na2SeSO3 in the solution increased, the cubic and snake-like CuSe nanostructures were formed gradually, the cubic nanostructures were captured by the CuSe nanosnakes, the CuSe nanosnakes grew wider and longer as the reaction time increased. Finally, the cubic CuSe nanostructures were completely replaced by BSA–CuSe nanosnakes. The prepared BSA–CuSe nanosnakes exhibited enhanced biocompatibility than the CuSe nanocrystals, which highly suggest that as-prepared BSA–CuSe nanosnakes have great potentials in applications such as biomedical engineering.
Keywords: Copper selenide, Nanosnakes, Bovine serum albumin, Synthesis, Characterization, Mechanism, Biocompatibility
Introduction
Copper selenides (CuSe) are well-known p-type semiconductors having potential applications in solar cells, optical filters, nanoswitches, thermoelectric and photoelectric transformers, and superconductors [1]. A lot of efforts have been devoted to the synthesis of copper selenides micro- and nanocrystallites with various morphologies, such as particles [2], tubes [3], cages [4], and flake-like structures [5]. There have been a few reports on the synthesis of copper selenide 1D nanomaterials. For example, Cu2−x Se nanowires with lengths of several micrometers and diameters of 30–50 nm have been prepared by employing selenium-bridged copper cluster as precursor in a chemical vapor deposition (CVD) process [6]. Also synthesized are arrays of copper selenide nanowires of mixed compositions of Cu3Se2/Cu2−x Se or Cu2−x Se/Cu in various proportions with lengths of several micrometers and diameters of 13–17 nm by using porous anodic aluminum oxide film as template [7]. However, to our knowledge, few reports are closely associated with the environmental-friendly controllable synthesis of 1D snake-like morphological CuSe nanomaterials based on biomolecule-assisted synthesis. For example, Muñoz-Rojas et al. [8] synthesized Ag@PPy nanomaterials that had snake-like shape and showed the properties of bending and folding under hydrothermal conditions while retaining the crystallographic coherence of the silver core, which were highly suggested that snake-like 1D nanomaterials might have some unique properties and potential application.
In recent years, biomimetic synthesis has become a hotspot [9]. For example, Yang et al. [10] reported biomimetic synthesis of Ag2S [10], HgS [11], and PbS [12], etc. in the bovine serum albumin (BSA) solution. These synthesized 1D nanomaterials have unique electrical, optoelectronic, biological, and mechanical properties with fundamental significance and great potential in applications such as electrochemical storage cells, solar cells, solid-state electrochemical sensors, semiconductive optical devices, catalyst, superionic materials, and biomedical engineering [13-16] and have attracted tremendous attentions from researchers in the field of materials, micro-electronics, and nanotechnology in recent years. However, how to fully use the advantage of bionanomaterials such as DNA, RNA and proteins, and metal nanomaterials as assistant media to fabricate 1D nanocomposites with controllable shapes and unique properties is still a great challenge. Up to date, few reports are associated with application of CuSe nanomaterials in biomedical engineering.
Herein, we selected one-dimensional copper selenide nanocrystals (CuSe) as research target, chose BSA as assistant reagent, developed a simple, nontoxic, room temperature, environmentally friendly method to synthesize controllably 1D BSA-wrapped copper selenide snake-like nanocomposites, and investigated these as-prepared products’ properties by UV–vis spectroscopy, high-resolution transmission electron microscopy, selected-area electron diffraction, energy dispersive spectroscopy, Raman spectroscopy, and MTT method. We found that as-prepared CuSe nanosnakes own some unique properties and enhanced biocompatibility, the possible formation mechanism of CuSe nanosnakes is also explored. Our primary results show that BSA–CuSe nanosnakes have great potential applications in biomedical engineering.
Experiments
Materials
All the reagents, including Cu(NO3)2, Na2SO3, and Se powder, were from Sinopharm Company, China. BSA with average molecular weight of about 68 KD was from Xiamen Sanland Chemicals Company Limited, China. All other reagents were from Sigma Inc. Human fibroblast cell line was obtained from the American Type Collection Company. RPMI 1640 medium containing 10% fetal calf serum was from Gibco Company. Agarose was from Sigma (St. Louis, United States). 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) was obtained from Dojin Laboratories (Kumamoto, Japan).
Synthesis of CuSe Nanosnakes
The Na2SeSO3 solution was prepared by refluxing selenium powder (5 mmol) and Na2SO3(5 mmol) in distilled water (200 ml) under nitrogen atmosphere for 24 h. In a typical synthesis process, 5 ml of 25 mM copper nitrate aqueous solution and 10 ml of 3 mg/ml BSA aqueous solution were mixed under vigorous stirring at room temperature (25°C). The mixed solution of the BSA–Cu2+ emulsion was kept static under nitrogen protection for 2 h. Then, 5 ml of 25 mM Na2SeSO3 solution was added. The color of the mixed solution rapidly changed to black. The mixed reaction solution was kept static under ambient conditions for 96 h, and then was separated by centrifugation at 15,000 rpm. The collected black solid-state products were washed with double distilled water and ethanol for three times and dried in a vacuum at room temperature for 24 h. During the process of nanosnakes growth, four replicas of the same experiment were run in parallel. Each replica was terminated at different times such as 24, 48, 72, and 96 h. To investigate the influence of BSA on the formation of copper selenide nanosnakes, a control experiment was carried out, copper selenide was prepared in the aqueous solution without BSA, and other conditions and procedures were the same as in a typical experiment.
Characterization of Synthesized BSA–CuSe Nanosnakes
These synthesized BSA–CuSe nanosnakes were characterized by a UNICAM UV300 spectrophotometer (Thermo Spectronic, USA), high-resolution transmission electron microscopy(HR-TEM, Hitachi H-700H, Hitachi, Japan), selected-area electron diffraction, energy dispersive spectroscopy, a PerkinElmer LS55 spectrofluorimeter, Laser Raman spectroscopy, and Fourier transform infrared (FT-IR) spectroscopy (an FTS135 infrared spectrometer from BIO-RAD, United States).
Cell Culture and MTT Analysis
Human fibroblast cell line was cultured in RPMI 1640, containing 1 × 105 mU/ml of penicillin and 0.1 mg/ml of streptomycin supplemented with 10% (v/v) FCS, at 37°C in a humidified 5% CO2 and 95% air atmosphere for 48 h. These cells were collected and added into 24-well plates at the concentration of 5,000 cells/well and continued to culture for 24 h. Then, the 100 μl CuSe nanocrystals (20 μg/ml) and 100 μl BSA–CuSe nanocrystals(20 μg/ml) were added into the 24-well plates, not added into the control wells, and continued to culture for 3 days. MTT (5 mg/ml) was prepared in PBS, and 20 μl was added to each well, and the cells were incubated for 4 h at 37°C, then the medium was removed, 200 μl dimethyl sulfoxide was added to each well, and optical density (OD) was read at 515 nm. The cell viability was calculated by the following formula: cell viability (%) = OD (optical density) of the treated cells/OD of the nontreated cells. The percentage of cell growth was calculated as a ratio of numbers of CuSe or BSA–CuSe nanosnakes-treated cells and control cells with 0.5% DMSO vehicle [17-19].
Statistical Analysis
Each experiment was repeated three times in duplicate. The results were presented as mean ± SD. Statistical significance was accepted at a level of P < 0.05.
Results and Discussion
Synthesis and Characterization of BSA–CuSe Nanosnakes
As shown in Fig. 1a, we can clearly observe the BSA–CuSe nanosnakes with different lengths. We also observed that the cubic copper selenide nanostructures were firstly formed in Fig. 1b. As the reaction time increased, the cubic copper selenide nanostructures gradually disappeared and became the nanosnakes. The resultant nanosnakes gradually grew longer and longer.
Figure 1.
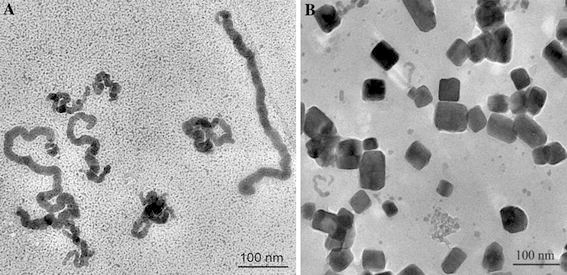
TEM images of BSA–CuSe nanosnakes (a) and the cubic copper selenide nanostructures (b)
Regarding the synthesis of one-dimensional BSA–CuSe nanostructures, sodium selenosulfate (Na2SeSO3) was used as Se source, which has been widely used to prepare nanocrystallite selenides such as CdSe [20] and PbSe [21]. Lakshmi et al. [22] and Nair et al. [23] have successfully prepared copper selenide (Cu2−xSe and Cu3Se2) thin films by using Na2SeSO3 as Se source. Na2SeSO3 is much more active than Se powder, because it reacts easily with Cu2+ ions at room temperature, is also less toxic, and, therefore, is safer to use than Na2Se or H2Se [24]. Equations (1) and (2) describe the reaction processes:
In the course of synthesis of 1D BSA–CuSe nanostructures, BSA was used as the soft-template to control the nucleation and growth of the nanocrystals, and also the dispersion and stabilization of the nanocrystals in solvents. As well known, BSA possesses a zwitterionic character at the isoelectric point (pI 4.7), displayed reversible conformational isomerization as the pH value changing [25]. BSA can bind with different sites of a variety of cationic and anionic groups, which makes possible utilization of BSA-decorated nanomaterials in a variety of supramolecular assemblies. For example, any conformational BSA can form covalent adduct with various metal ions [26], such as Cu2+, Ni2+, Hg2+, Ag+, and AuCl4−.
Potential Mechanism of BSA–CuSe Nanosnake Formation
In order to clarify the mechanism of synthesis of CuSe nanosnakes, we characterized the CuSe nanostructures by UV–vis spectroscopy. Figure 2a shows the UV–vis absorption spectra of pure BSA, BSA–Cu2+, and BSA–CuSe. The pure BSA has a special absorption peak at 280 nm. The spectrum of BSA–Cu2+ complex did not display shift and enhancement of absorption peak at 280 nm, because the BSA protein can provide multiple binding sites for Cu2+. The spectrum of BSA–CuSe nanocomposites clearly showed the absorption peak shift from 280 nm to 228 nm after the Na2SeSO3 solution was added into the BSA–Cu2+ solution, indicating that the Se2− released from Na2SeSO3 and reacted with Cu2+ forming CuSe nanostructures. Figure 2b clearly shows that the absorbance of BSA–CuSe nanostructures was markedly enhanced, because more and more BSA–CuSe nanosnakes were formed.
Figure 2.
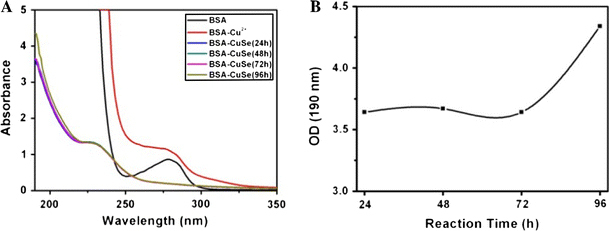
a UV–vis absorption spectrum of BSA, BSA–Cu2+, BSA–CuSe at 24 h, BSA–CuSe at 48 h, BSA–CuSe at 72 h, BSA–CuSe at 96 h; b The change of absorbance at 190 nm
The BSA–CuSe nanosnake was also characterized by using high-resolution transmission electron microscopy (HRTEM), selected-area electron diffraction (SAED), and energy dispersive spectroscopy (EDS). Figure 3a–3c show representative TEM images of the BSA–CuSe nanosnakes at the different reaction time such as 24, 48, and 96 h, respectively. We can clearly observe that the well-dispersed nanostructures displayed different sizes, representing the different growth stages. Within the 24-h reaction time, BSA–CuSe nanostructures mainly exhibited cubic structure with average size of 30 nm. After 24 h, the BSA–CuSe nanosnakes formed gradually, their sizes were about 130 nm in length and 12 nm in width. After 48 h, the cubic nanostructures had little change, short rods appeared, and the nanosnakes grew wide and long (Fig. 3b). When the reaction time reached to 96 h, the cubic nanostructures almost completely disappeared, and the nanosnakes grew homogeneously up to about 200 nm in length, and 14 nm in width (Fig. 3c, 3f). When the reaction time was over 96 h, the sizes of nanosnakes were almost unchanged. As shown in Fig. 3d, the single nanosnake exhibits good crystalline and clear lattice fringes. The lattice fringe spacing was 0.172 nm, consistent with the interplanar spacing of the (113) plane of cubic berzelianite (Cu2−xSe) crystallites. Figure 3e is the corresponding SAED pattern, revealing that the nanosnakes are crystalline and can be indexed to berzelianite Cu2−xSe.
Figure 3.
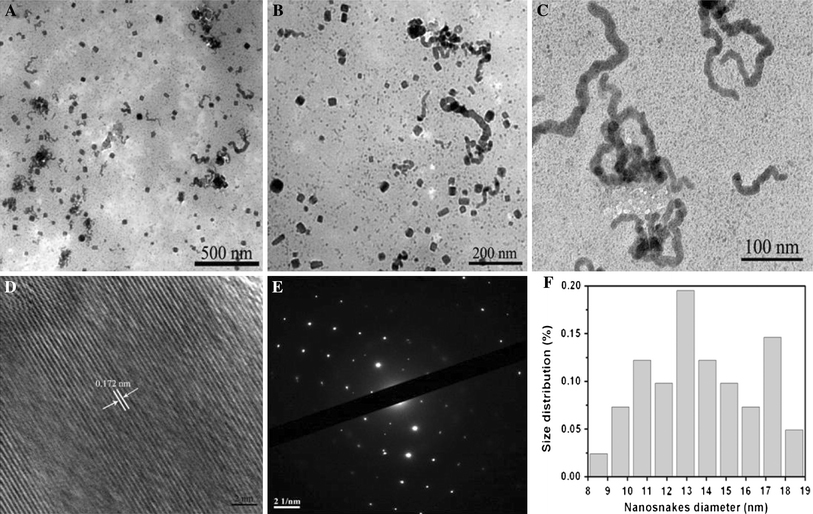
TEM images of BSA–CuSe nanosnakes obtained after different aging time in the typical experiment: a 24 h, b 48 h, and c 96 h, respectively. d HRTEM image of an individual nanosnake. e SAED pattern in an area including many nanosnakes. f The histogram of nanosnakes at 96 h
In order to investigate the typical growth stage of nanosnakes, we used HR-TEM to observe the samples at 48-h reaction time. Figure 4a depicts the typical morphology of the cubic BSA–CuSe nanocomposites revealing that the peanut-like assemblies and shorter nanorods were generated. As shown in Fig. 4b, the adjacent nanostructures attached without sharing a same crystallographic orientation. The experimental lattice fringe spacing, 0.146 nm, is consistent with the interplanar spacing in monoclinic Cu2Se. The connected nanoparticles were rotated to find the common crystallographic orientation (indicated by the white arrow) [27]. After the rotations were finished, they fused to form almost a perfect short nanorod (Fig. 4c). The lattice fringe spacing is 0.173 nm, which was in agreement with the interplanar spacing of the (113) plane of cubic berzelianite (Cu2−xSe) crystallites.
Figure 4.
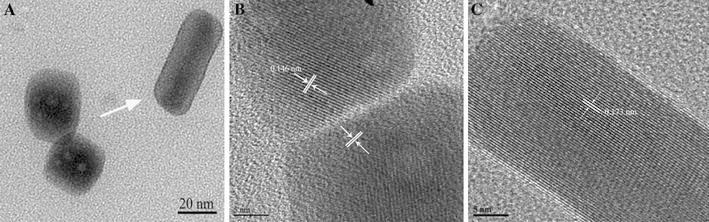
TEM images showing oriented attachment of cubic copper selenide in BSA solution for 48 h. a Low-magnification TEM image of sample. b HRTEM image of two primary crystallites forming “peanut” or “chain” via oriented attachment. c HRTEM image of a single nanorod after being fused together
To understand the growth mechanism of nanosnakes, the representative TEM images of the devourment of cubic nanoparticles were recorded in Fig. 5a–5c, and the prepared nanosnakes also characterized by scanning electron microscope (SEM) (see Supplementary Fig. 2). When the cubic nanoparticles were captured by the nanosnakes, the square boundary gradually fuzzed with the reaction time and disappeared finally (shown by the white arrow in Fig. 5a). Figure 5b, 5c showed the capture transient and the devourment stage, respectively. Figure 5d–5f recorded three different parts of an individual nanosnake, including the neck (D), the body (E), and the tail (F), whose lattice fringe spacing is respectively 0.266, 0.101, and 0.155 nm. The different parts have different crystallographic orientation and steadily existed in BSA solution. The EDS spectrum shows the presence of elements Cu and Se in the prepared nanosnakes (see Supplementary Fig. 3). The peaks of C and O element are due to the BSA. The Supplementary Table 1 documents the weight percentage and atomic percentage of silver and selenium elements of the measured area, which showed that the atomic ratio of Cu and Se does not match the stoichiometric molar ratio (Cu/Se) of copper selenide exactly. The main reason is that the amount of selenium in the BSA solution is excessive (see Supplementary Table 1). According to the above phenomena, it could be presumed that the nanosnakes are growing at the expense of the colloidal particles in the Ostwald ripening process, and BSA act as a stabilizing agent to modify the new generated nanosnakes surface.
Figure 5.
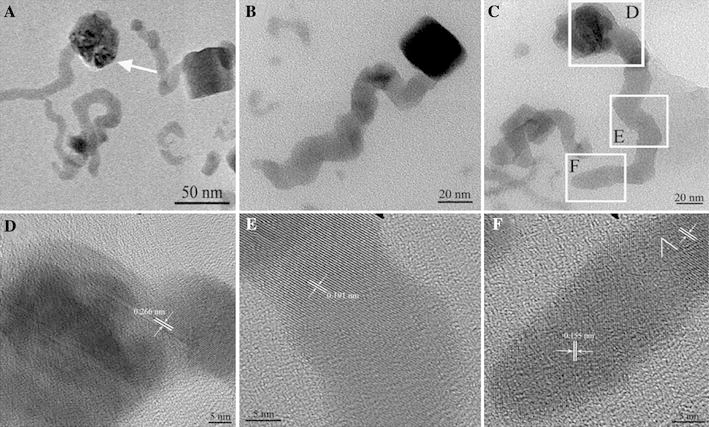
TEM images showing oriented attachment of copper selenide nanosnakes in BSA solution for 48 h. a Low-magnification TEM image of sample. b, c TEM images of two different devour stages of copper selenide nanosnakes. HRTEM images of different parts of an individual nanosnake: d the neck, e the body, f the tail, respectively
To clarify the formation mechanism of BSA–CuSe nanosnakes, we also obtained the FT-IR spectra and Raman spectra of pure BSA, BSA–Cu2+, and BSA–CuSe powders. The FT-IR spectra and the data of the main peaks are shown in Fig. 6a and supplementary Table 2. The IR peaks of pure BSA at 3,430, 3,062, 1,652, and 1,531 cm−1 are assigned to the stretching vibration of –OH, amide A (mainly—NH stretching vibration), amide I (mainly C=O stretching vibrations), and amide II (the coupling of bending vibrate of N–H and stretching vibrate of C–N) bands, respectively. The difference between the IR spectrum of pure BSA and that of BSA–Cu2+ is obvious. The characteristic peak of –NH groups disappeared, suggesting that there might be coordination interaction between Cu2+ and –NH groups of BSA, which may play an important role in the formation of CuSe nanosnakes. In addition, the new peaks of BSA–Cu2+ at 1,021 and 824 cm−1 might be contributed to the interaction of Cu2+ and BSA. The strong peak at 1,383 cm−1 in the BSA–Cu2+, and BSA–CuSe spectra is attribute to the absorption of NO3−1, which was introduced by the addition of Cu(NO3)2.Comparing the IR spectra of BSA–CuSe with those of pure BSA, the characteristic peak of –OH groups shifts to a high wavenumber of about 5 cm−1, and the characteristic peak of –NH groups disappears. The results indicate that the conjugate bonds existed between the CuSe nanosnakes and –OH groups and –NH groups of BSA.
Figure 6.
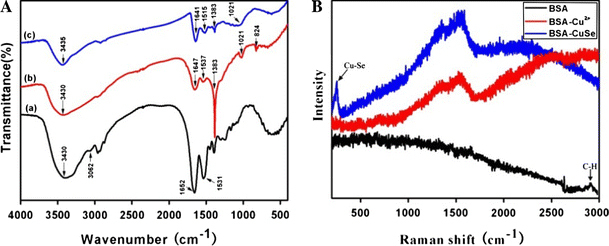
a The FT-IR spectra of (a) pure BSA, (b) BSA–Cu2+, and (c) BSA–CuSe in BSA solution for 96 h. b Raman spectra (632.8 nm excitation) of pure BSA, BSA–Cu2+, BSA–CuSe in BSA solution for 96 h
Raman spectroscopy is used to investigate the changes in the electronic properties of nanomaterials through the special electron–phonon coupling that occurs under strong resonant conditions. Therefore, Raman spectra are very powerful to detect of the new chemical bonds. As shown in Fig. 6b, the difference between the Raman spectrum of pure BSA and that of BSA–Cu2+ is obvious. The bands C–H of BSA at 2,926 cm−1 disappeared, suggesting that there might be coordination interaction between Cu2+ and BSA. Comparing the Raman spectra of BSA–CuSe with those of pure BSA and BSA–Cu2+, the characteristic peak of Cu–Se bonds at 250 cm−1 was found, which is consistent with the standard Raman spectra of cubic berzelianite (Cu2−xSe) crystallites(RRUFF ID: R060260.2). The above facts highly suggested that the Cu2−xSe nanosnakes were successfully synthesized in the BSA solution.
To further study the formation mechanism of the nanosnakes in the BSA aqueous solution, the conformation changes in the secondary structures of BSA in the reaction system were determined by CD spectroscopy, which is a valuable spectroscopic technique for studying protein and its complex. The CD spectra of pure BSA, BSA–Cu2+, and BSA–CuSe solutions are shown in Fig. 7. From the figure, it can be seen that the CD curve of BSA–Cu2+ solution is similar to that of the pure BSA solution, while the CD spectrum of the BSA–CuSe solution is different from that of pure BSA. According to the result, it can be seen that copper ions only induced the smaller deformation of the BSA molecules in the BSA–Cu2+ solution, whereas there were bigger changes in the BSA conformation in the BSA–CuSe nanosnake solution, resulting from the strong conjugate bonds between BSA and surfaces of the colloidal nanosnakes. With the growth of CuSe nanosnakes, more and more α-Helix were stretched and transformed into β-Sheets, which could be contribution to the impairment or break of hydrogen bonds.
Figure 7.
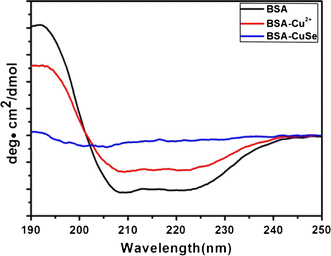
The CD spectra of a pure BSA, b BSA–Cu2+, and c BSA–CuSe in BSA solution
According to the data mentioned above, we suggest one possible mechanism model of CuSe nanosnake formation based on use of BSA as soft-template, shown in Scheme 1. The basic principle is attributed to that whose structure decides whose function. BSA has reversible conformational isomerization in different pH condition, when pH value is lower than 4.7, BSA undergoes another expansion with a loss of the intra-domain helices (10) of domain I which is connected to helix (1) of domain II and that of helix (10) of domain II connected to helix (1) of domain III [27,28] Then, BSA has three reversible forms: N forms, F forms, and E forms, which could bind with CuSe nanoparticles, finally result in the formation of different shapes of CuSe nanostructures, for example, CuSe nanoparticles bound with N forms formed the sphere nanostructures, CuSe nanoparticles bound with F forms formed the cubic nanostrctures, and CuSe nanoparticles bound with E forms formed the nanosnakes, final CuSe nanostrcutures strongly depend on the structures of BSA proteins under the reaction condition.
Scheme 1.

Schematic mechanism of synthesis of CuSe nanosnakes using BSA as soft-template
Biocompatibility of CuSe Nanocrystals and BSA–CuSe Nanosnakes
As shown in Fig. 8, as the culture days increased, the cell viability decreased accordingly, the cell viability in BSA–CuSe group was markedly higher than that in CuSe nanocrystals group, there existed statistical difference between two groups (P < 0.01), which shows that BSA–CuSe nanosnakes own better biocompatibility than the CuSe nanocrystals.
Figure 8.
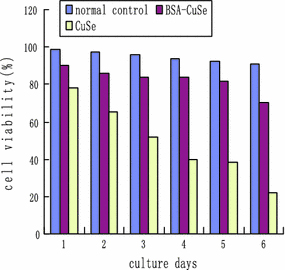
Effects of 20 μg/ml BSA–CuSe nanosnakes and CuSe nanocrystals on Human fibroblast cells
Conclusions
In conclusion, CuSe nanosnakes were successfully synthesized at room temperature using BSA as soft-template. Regarding the potential mechanism of the phenomena, we suggested a possible model: at first, the cationic Cu2+ions were covalently adducted to BSA, as the amounts of selenide ions (Se2−) released from Na2SeSO3 in the solution increased, the cubic and snake-like CuSe nanostructures were formed gradually. Secondly, the cubic nanostructures were captured by the CuSe nanosnakes, the CuSe nanosnakes grew wider and longer as the reaction time increased. Finally, the cubic CuSe nanostructures were completely replaced by BSA–CuSe nanosnakes. The prepared BSA–CuSe nanosnakes exhibited enhanced biocompatibility than the CuSe nanocrystals, which highly suggest that as-prepared BSA–CuSe nanosnakes have great potentials in applications such as biomedical engineering.
(See supplementary material 1)
Supplementary Material
Supplementary Figures 1-3 and Table.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No.20803040 and No.20471599), Chinese 973 Project (2010CB933901), 863 Key Project (2007AA022004), New Century Excellent Talent of Ministry of Education of China (NCET-08-0350), Special Infection Diseases Key Project of China (2009ZX10004-311), Shanghai Science and Technology Fund (10XD1406100). The authors thank the Instrumental Analysis Center of Shanghai Jiao Tong University for the Materials Characterization.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Bhuse VM, Hankare PP, Garadkar KM, Khomane AS. Mater. 2003. p. 82. COI number [1:CAS:528:DC%2BD3sXhs1aktLc%3D] [DOI]
- Zhang WX, Zhang XM, Zhang L, Wu JX, Hui ZH, Cheng YW, Liu JW, Xie Y, Qian YT. Inorg. 2000. p. 1838. COI number [1:CAS:528:DC%2BD3cXitF2gs78%3D] [DOI] [PubMed]
- Xu J, Zhang WX, Yang ZH, Yang SH. Inorg. 2008. p. 699. COI number [1:CAS:528:DC%2BD2sXhsVamsbvP] [DOI] [PubMed]
- Cao HL, Qian XF, Zai JT, Yin J, Zhu ZK. Chem. 2006. doi: [DOI] [PubMed]
- Xie Y, Zheng XW, Jiang XC, Lu J, Zhu LY. Inorg. 2002. p. 387. COI number [1:CAS:528:DC%2BD3MXptlSqtbc%3D] [DOI] [PubMed]
- Hsu YJ, Jiang CM, Lin YF, Liaw BJ, Lobana TS, Lu SY, Liu CW. Chem. 2006. p. 3323. COI number [1:CAS:528:DC%2BD28XlsFSiuro%3D] [DOI]
- Jagminas A, Juškėnas R, Gailiūtė I, Stakutė G, Tomašiūnas R. J. Cryst. Growth. 2006. p. 343. COI number [1:CAS:528:DC%2BD28XptVCmsrw%3D]; Bibcode number [2006JCrGr.294..343J] [DOI]
- Muñoz-Rojas D, Oró-Solé J, Gómez-Romero P. J. Phys. Chem. C. 2008. p. 20312. COI number [1:CAS:528:DC%2BD1cXhsVKis7nJ] [DOI]
- Yu S. Top Curr. 2007. p. 79. COI number [1:CAS:528:DC%2BD2sXhsFers7s%3D] [DOI]
- Yang L, Xing R, Shen Q, Jiang K, Ye F, Wang J, Ren Q. J. Phys. Chem. B. 2006. p. 10534. COI number [1:CAS:528:DC%2BD28XktFeitrk%3D] [DOI] [PubMed]
- Qin D, Ma X, Yang L, Zhang L, Ma Z, Zhang J. J. 2008. p. 559. COI number [1:CAS:528:DC%2BD1cXisFCqtr0%3D] [DOI]
- Zhang J, Ma X, Guo Y, Yang L, Shen Q, Wang H, Ma Z. Mater. 2010. p. 112. COI number [1:CAS:528:DC%2BD1MXhtlGgtrnK] [DOI]
- Wen X, Wang S, Xie Y, Li X, Yang S. J. Phys. Chem. B. 2005. p. 10100. COI number [1:CAS:528:DC%2BD2MXjsFahtLw%3D] [DOI] [PubMed]
- Goldberger J, He R, Zhang Y, Lee S, Yan H, Choi H, Yang P. Nature. 2003. p. 599. COI number [1:CAS:528:DC%2BD3sXislCgtb4%3D]; Bibcode number [2003Natur.422..599G] [DOI] [PubMed]
- He C, Zhang L, Wang H, Zhang F, Mo X. Nano Biomed. 2009. p. 119. COI number [1:CAS:528:DC%2BC3cXksFensrg%3D] [DOI]
- Chen F, Huang P, Mo X. Nano Biomed. 2010. p. 84. COI number [1:CAS:528:DC%2BC3cXksFens70%3D]; Bibcode number [2009nane.conf...84C]
- Cui D, Tian F, Ozkan CS, Wang M, Gao H. Toxicol. 2005. p. 73. COI number [1:CAS:528:DC%2BD2cXhtVCktrjP] [DOI] [PubMed]
- Wang Z, Ruan J, Cui D. Nanoscale Res. 2009. p. 593. COI number [1:CAS:528:DC%2BD1MXnt1Sktrw%3D]; Bibcode number [2009NRL.....4..593W] [DOI] [PMC free article] [PubMed]
- Pan B, Cui D, Xu P, Ozkan CS, Gao F, Ozkan M, Huang T, Chu B, Li Q, He R, Hu G. Nanotechnology. 2009. p. 125101. COI number [1:CAS:528:DC%2BD1MXltFCns78%3D]; Bibcode number [2009Nanot..20l5101P] [DOI] [PubMed]
- Kale S, Lokhande C. Mater. 2000. p. 103. COI number [1:CAS:528:DyaK1MXnvFaksr8%3D] [DOI]
- Kitaev G, Khvorenkova A. Russ. 1998. p. 1325.
- Lakshmi M, Bindu K, Bini S, Vijayakumar K, Kartha C, Abe T, Kashiwaba Y. Thin Solid Films. 2001. p. 127. COI number [1:CAS:528:DC%2BD3MXhvVajsr8%3D]; Bibcode number [2001TSF...386..127L] [DOI]
- García V, Nair P, Nair M. J. Cryst. Growth. 1999. p. 113. Bibcode number [1999JCrGr.203..113G] [DOI]
- Ge J, Xu S, Liu L, Li Y. Chem. 2006. p. 3672. COI number [1:CAS:528:DC%2BD28XksV2iu7c%3D] [DOI] [PubMed]
- Carter D, Ho J. Adv. 1994. p. 153. COI number [1:CAS:528:DyaK2cXltVajtLk%3D] [DOI] [PubMed]
- Friedli G. PhD Dissertation. University of Surrey, Guildford; 1996. [Google Scholar]
- Yang L, Yang H, Yang Z, Cao Y, Ma X, Lu Z, Zheng Z. J. Phys. Chem. B. 2008. p. 9795. COI number [1:CAS:528:DC%2BD1cXosl2jtrc%3D] [DOI] [PubMed]
- Xu J, Zhang W, Yang Z, Ding S, Zeng C, Chen L, Wang Q, Yang S. Adv. 2009. p. 1759. COI number [1:CAS:528:DC%2BD1MXnslCksrc%3D] [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-3 and Table.


