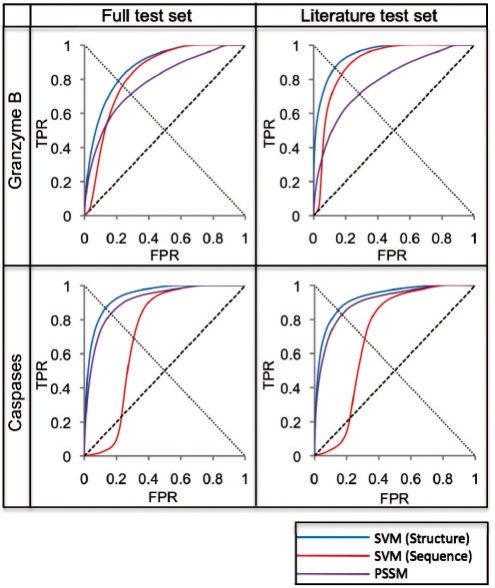
Fig. 3.
Benchmark results. Results from different methods applied to four different datasets, represented by ROC curves. The line from (0,0) to (1,1) represents a random predictor; a perfect classifier would go from (0,0) to (0,1) and then to (1,1). The critical point of the ROC curve is where each curve intersects the line from (1,0) to (0,1). Full test sets included all known substrates for the respective protease type, and Literature test sets excluded the large proteomic datasets, retaining only the GrBah and Casbah substrates. SVM (Structure) was developed in the current study; SVM (Sequence) was taken from a previous study that trained on cleavage sequence residue type only (Wee et al., 2006); PSSM implemented the GrabCas method for GrB substrates (Backes et al., 2005), while for caspases it was trained on frequency of residue types at each position in known cleavage sequences, using the PoPS (Boyd et al., 2005) algorithm. All ROC plots were interpolated through a number of points equal to the number of test set positives in each dataset (Supplementary Fig. 1a).