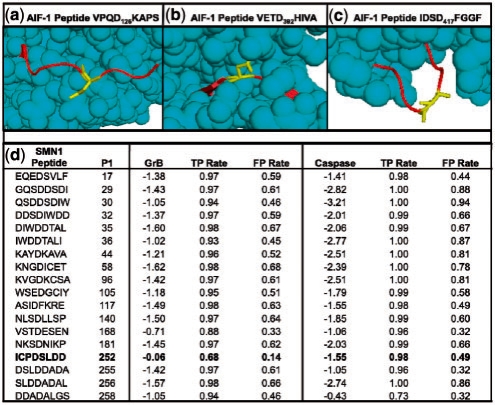
Fig. 5.
Details of novel GrB substrates. (a) Solved structure of AIF-1 (PDB ID 1M6I), highlighting Asp126. Cleavage at this site is consistent with the observed banding patterns on the immunoblot. (b) A peptide on AIF-1 centered on Asp392 that scores well when examining sequence only, but poorly when structure is considered, likely due to being largely inaccessible to solvent. (c) Another high scoring site on AIF-1 at Asp417; it is unclear why this site is not cleaved despite favorable sequence and structure properties. (d) Scores for the SMN1 protein for all octopeptides with Asp in the fourth position, as assessed by SVMs trained on GrB and caspase substrates, respectively. The ‘GrB’ and ‘Caspase’ columns indicate the scores that the respective SVMs assigned to each peptide, and TPR and FPR signify rates that these scores would fall on in the benchmark set.