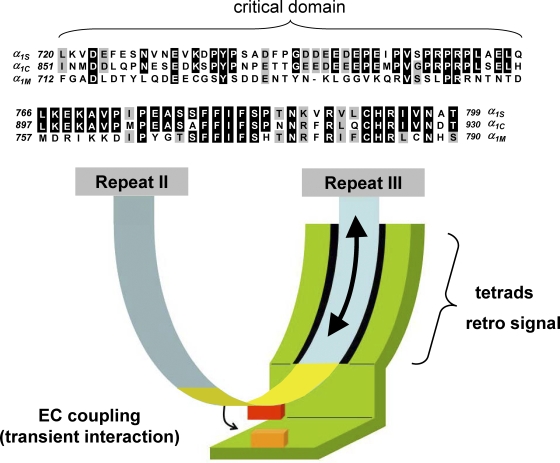
Figure 1.
Model for the engagement of skeletal muscle EC coupling based on the current literature. (Top) Sequence alignment is shown for rabbit α1S, rabbit α1C, and Musca domestica α1M. The top panel (α1S residues 720–765) represents the critical domain (Beam and Horowicz, 2004). The middle panel (α1S residues 766–799) represents the region of the α1S II-III loop carboxyl-terminal to the critical domain, ending just before repeat III. Sequence is not shown for the relatively divergent amino-terminal portion of the II-III loop (α1S residues 662–719). Residues of α1C or α1M identical to those of α1S are shown boxed in black, and residues conserved with those of α1S are shown boxed in gray. (Bottom) The arc extending from repeat II to repeat III represents the α1S II-III loop; the yellow portion of the loop represents the critical domain (α1S residues 720–765). The green entity represents the junctional interaction partner(s) of the α1S II-III loop. Because the amino-terminal portion of the α1S II-III loop is accessible to large streptavidin probes, it is depicted as being devoid of junctional interaction partners. Because ultrastructural analysis of α1S/α1M chimeras suggests that the carboxyl-terminal portion of the α1S II-III loop supports junctional contacts that are essential for tetrad formation, the carboxyl-terminal portion of the loop is shown as a surface for interaction with other junctional proteins. In addition, the carboxyl-terminal portion of the loop may also serve as a line of communication (large arrow) between repeat III and the critical domain (yellow) in the center of the α1S II-III loop; voltage-induced conformational rearrangements (little arrow) in the critical domain engage SR Ca2+ release via a transient protein–protein interaction between a portion of the critical domain (red box) and another junctional protein (orange box).