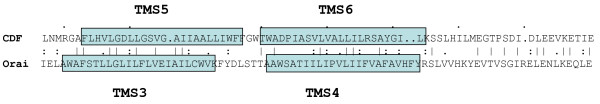
Figure 4.
Alignment of the Consensus Sequence for the CDF Family (TMSs 5 and 6 and flanking regions) with the Consensus Sequence for the Orai Family (TMSs 3 and 4 and flanking regions). The consensus sequences were generated using the HMMER package (see Methods). The two consensus sequences were aligned using the GAP program (GCG package). This alignment shows 30% identity and 46% similarity with 3 gaps. TMSs were predicted with the TMHMM program http://www.cbs.dtu.dk/services/TMHMM. Each amino acid shown in the consensus sequence is the highest probability amino acid at that position according to the Hidden Markov Model.