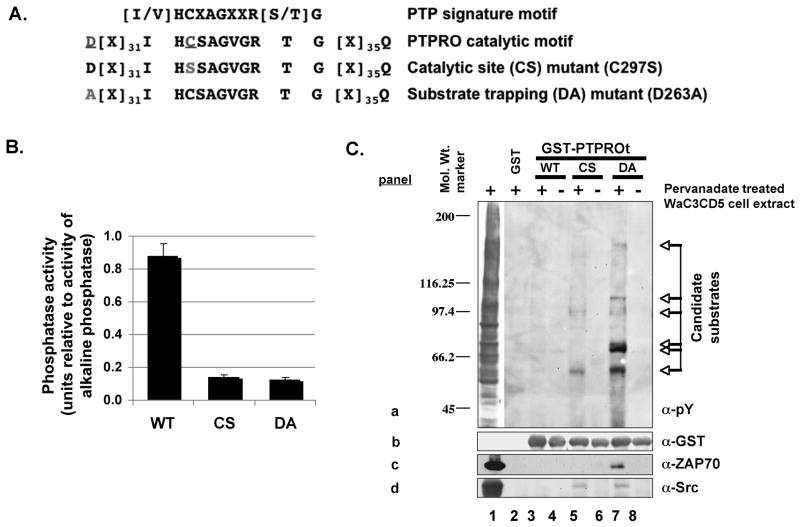
Fig. 1. Substrate-trapping assay identified several candidate pY-polypeptides as PTPROt substrate.
(A) Schematic representation of the catalytic site and substrate-trapping mutants of PTPROt used in this study. (B) Activity of PTPROt (WT and mutants) measured using pNPP as substrate. Equal amounts of GST-tagged PTPROt purified using GSH-sepharose was assayed for activity in parallel with increasing concentration of alkaline phosphatase. A plot of units of alkaline phosphatase v/s optical density of product p-nitrophenol was used to compute the activity of PTPROt. (C) Whole cell extract of pervanadate treated B-cell line WaC3CD5 was used to bind bacterially expressed and purified GST-tagged PTPROt (WT, CS, DA). The pulled-down proteins separated by SDS-PAGE were immonoblotted with anti-pY cocktail (4G10, pY20, pY99) (panel a), anti-GST (panel b), anti-ZAP70 (panel c) or anti-Src (panel d). The positions of molecular weight standards (in kDa) are indicated on the left and candidate substrates are denoted by arrows on the right.