Abstract
Platelet-derived growth factors (PDGFs) and their receptors (PDGFRs) are prototypic growth factors and receptor tyrosine kinases which have critical functions in development. We show that PDGFs share a conserved region in their prodomain sequences which can remain noncovalently associated with the mature cystine-knot growth factor domain after processing. The structure of the PDGF-A/propeptide complex reveals this conserved, hydrophobic association mode. We also present the structure of the complex between PDGF-B and the first three Ig domains of PDGFRβ, showing that two PDGF-B protomers clamp PDGFRβ at their dimerization seam. The PDGF-B:PDGFRβ interface is predominantly hydrophobic, and PDGFRs and the PDGF propeptides occupy overlapping positions on mature PDGFs, rationalizing the need of propeptides by PDGFs to cover functionally important hydrophobic surfaces during secretion. A large-scale structural organization and rearrangement is observed for PDGF-B upon receptor binding, in which the PDGF-B L1 loop, disordered in the structure of the free form, adopts a highly specific conformation to form hydrophobic interactions with the third Ig domain of PDGFRβ. Calorimetric data also shows that the membrane-proximal homotypic PDGFRα interaction, albeit required for activation, contributes negatively to ligand binding. The structural and biochemical data together offer insights into PDGF-PDGFR signaling, as well as strategies for PDGF-antagonism.
Keywords: crystallography, receptor tyrosine kinase, signal transduction
Platelet-derived growth factors (PDGF-A, -B, -C, and -D) are major mitogens for connective tissue cells such as fibroblasts and smooth muscle cells (1, 2), and critically regulate embryonic development (reviewed in ref. 3). The two receptors for PDGFs are PDGFRα and PDGFRβ. PDGFRα signaling controls gastrulation and the development of many organs, including lung, intestine, skin, testis, kidney, skeleton, and neuroprotective tissues, whereas PDGFRβ signaling has been well established in early hematopoiesis and blood vessel formation (3). While being essential for establishing specific developmental stages, PDGF-PDGFR signaling in adulthood is usually detrimental. Enhanced PDGF-PDGFR signaling is a hallmark in a variety of diseases, including cancers, atherosclerosis, pulmonary fibrosis, and restenosis (4).
PDGFs, and their relatives VEGFs, function as disulfide-linked dimers. They have an evolutionarily conserved cystine-knot-fold growth factor domain of ∼100 amino acids (aa), denoted the PDGF/VEGF homology domain, involved in receptor-binding and dimerization (5). Biosynthesis and processing of PDGFs are highly regulated. All PDGFs contain N-terminal prodomains of various length (70, 65, 212, 239aa for PDGF-A, -B, -C, and –D respectively; PDGF-C and –D have a Complement proteins C1r/C1s, Uegf, Bmp1 (CUB) domain in this region), which are proteolytically cleaved for activation when secreted (5). The molecular roles of these prodomains are not completely understood. PDGFRs belong to the class III receptor tyrosine kinases (RTKs), a five-member clan that includes PDGFRα, PDGFRβ, KIT, FMS, and FLT3, which are characterized by a 5-Ig-domain extracellular segment (designated D1–D5) and a split intracellular kinase domain (6), and bind ligands of either the cystine-knot-β-sheet fold or the four-helix-bundle fold. The dimerization and activation of KIT and FMS by their four-helix-bundle ligands have been well studied structurally (7–9), but a structure demonstrating how PDGFRs are dimerized and activated by cystine-knot-β-sheet ligands has been lacking. Since PDGFs and VEGFs are of common origin (10), and PDGFRs (with 5 Ig domains) and VEGFRs (with 7 Ig domains) are also related (6), the core of their ligand/receptor recognition complexes is perceived to be similar. Shedding light on this, the structure of VEGF-C in complex with the second and third domains of VEGFR2 has recently been determined (11). The four PDGFs bind PDGFRs with different specificity. Although functional evidence only exists for the homodimeric PDGF-A/PDGFRα, PDGF-C/PDGFRα, and PDGF-B/PDGFRβ complexes, biochemical data support additional homodimeric and heterodimeric combinations (5). The basis for such specificity is unclear.
In recent years, inhibiting PDGF-PDGFR signaling has become an attractive pursuit in anticancer therapy (4), and combined inhibition of PDGF and VEGF has emerged as a promising strategy for suppressing angiogenesis in tumor progression (12, 13). Several strategies have been utilized to block PDGF/PDGFR signaling at the extracellular level: neutralizing antibodies for PDGF ligands and receptors, aptamers, N-terminal processing-deficient PDGFs, and soluble receptors without the kinase domain (3). The development of such therapies would greatly benefit from a detailed structural model of PDGF/PDGFR interaction.
In this study, we have determined the crystal structures of a complex between PDGF-A and its propeptide, and a complex between PDGF-B and the first three extracellular domains of its receptor PDGFRβ. These structures reveal the conserved mode of PDGF:propeptide association, and the conserved mode of PDGF:PDGFR recognition. Our data offer unique insights into PDGF-PDGFR signaling, and provide a much needed template for designing anticancer therapies.
Results and Discussion
PDGFs Have Retained Propeptides During Recombinant Expression.
To express PDGFs, we cloned the receptor-binding domains of PDGFs (-A, -B, -C, and –D), with and without the prosequences, into the baculovirus-mediated mammalian gene transduction (BacMam) expression vector (14), and added a C-terminal His-tag to capture the secreted proteins. With the exception of PDGF-C, the PDGFs could not be secreted without the prosequences, indicating that they require prodomains for correct folding, consistent with previous data (reviewed in refs. 15, 16). The PDGFs were only partially processed by furin or furin-like proprotein convertases (PCs), as evidenced by the existence of both uncleaved and cleaved species in the affinity-purified proteins (Fig. S1). The prosequences are located N-terminal to the mature sequences, therefore we expected that the propeptides of PDGFs, once cleaved, would be undetectable following C-terminal affinity purification. To our surprise, the propeptides of PDGF-A, -B, and –D were all retained during purification of the mature peptides (Fig. S1 A and B), indicating that the propeptides were noncovalently associated with the mature peptides. In the cases of PDGF-A and PDGF-B, the propeptides are small (65–70 aa, ∼8 kDa), and were often detected as a diffuse band at the front line in SDS-PAGE analysis. When the “RRKR” furin-recoginition sequence (residues 83–86) of pro-PDGF-A was mutated to “QQQQ,” both the cleaved mature PDGF-A and the small peptide were no longer observed during purification (Fig. S1A, right gel). For PDGF-D, we observed that the CUB domain-containing prosequence (∼230 aa) cleaved from mature peptide, which was likely processed at the RKSK sequence (residues 254–257), was retained during metal affinity purification. However, a peptide smaller than the entire proregion persisted at the front edge in SDS-PAGE (Fig. S1B), suggesting a possible second cleavage site upstream of the RKSK sequence. That PDGF-A, -B, and –D all contain a < 100 aa prosequence noncovalently associated with the mature peptide suggests a possibility that there is a conserved mode of interaction between propeptides and mature PDGFs (discussed below).
The Structure of the PDGF-A/Propeptide Complex.
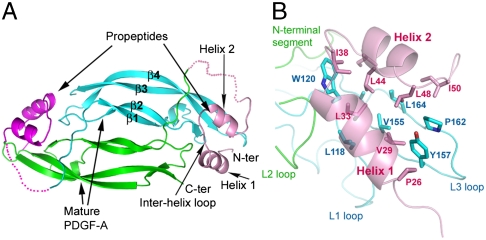
To probe the PDGF-propeptide interaction mode, we crystallized propeptide-bound PDGF-A, and determined its structure using molecular replacement (SI Text, Table S1, and Fig. 1). The PDGF-A dimer in the PDGF-A/propeptide complex is similar to the free PDGF-B structure in the core scaffold (17), with each protomer being a juxtaposition of two long, highly twisted antiparallel β-sheets of four strands (β1, β2, β3, and β4), joined by three loops (L1, L2, and L3). At the ends of juxtaposed β-sheets, two clamps are formed by the loops and the C-terminal segments. Each clamp has two unequal arms: the longer arm formed by the L1 and L3 loops from one protomer, designated the “protruding protomer;” and the shorter arm formed by the L2 loop and C terminus from the other protomer, designated the “receding protomer.” A large, hydrophobic groove is formed between the two protomers at each clamp.
Fig. 1.
Structure of the PDGF-A/propeptide complex. (A) Ribbons model of the complex, with the mature PDGF-A protomers in green and blue, and the propeptides in pink and magenta. (B) The hydrophobic PDGF-A/propeptide interface. The side chains involved in the PDGF-A/propeptide interaction are depicted as sticks.
The core of the propeptide is a juxtaposition of two short helices (residues 26–36 for helix 1 and residues 41–48 for helix 2) linked by a short loop (residues 37–40) (Fig. 1A). The N-terminal segment preceding helix 1 (residues 21–25) is well ordered, but the segment after helix 2, leading to the N terminus of mature PDGF-A, is disordered after residue 53. The flexibility of this segment preceding the PC-recognition site may facilitate the presentation of the substrate sequence (RRKR) to PC.
The PDGF-A/propeptide interaction features a flat attachment of the propeptide helices to the upper face of the mature PDGF-A clamps, near the N terminus of the mature domain. The interface is extensive and primarily hydrophobic, burying ∼2,070 Å2 solvent-accessible surface area. The two propeptide helices are amphipathic, utilizing their hydrophobic sides to interact with the large hydrophobic area on mature PDGF-A, and exposing their hydrophilic sides. The hydrophobic surface of the propeptide consists of Pro26, Val29, Leu33, and Ala34 from helix 1; Ile38 from the interhelix loop; Ile41, Leu44, and Leu48 from helix 2; and Ile50 and Leu53 from the posthelix 2 segment (Fig. 1B). This surface serves to cover the large hydrophobic cluster on the protruding PDGF-A protomer, consisting of Leu118 and Trp120 from the L1 loop; and Ala153, Val155, Tyr157, Pro162, and Leu164 from the L3 loop (Fig. 1B). The receding PDGF-A protomer also contributes to propeptide-binding, mostly through its L2 loop (Table S2).
Sequence alignment of the PDGF sequences preceding the growth factor domains indicates that the hydrophobic residues central to the PDGF-A/propeptide interaction are conserved among PDGFs (Fig. S1D). Therefore the PDGF-A/propeptide interaction mode is likely shared by other PDGFs.
The Structure of the PDGF-B/PDGFRβ-D1-D3 Complex.
To prepare the PDGF-PDGFR complexes, we mixed mammalian-cell-expressed PDGFRα and PDGFRβ extracellular segments with the appropriate PDGFs, and succeeded in crystallizing the PDGF-B/PDGFRβ-D1-D3 complex. The structure of the complex was solved by molecular replacement (SI Text and Fig. S2). The asymmetric unit contains one dimeric PDGF-B:PDGFRβ complex. The refined model consists of residues 1–102 for each PDGF-B protomer and residues 33–312 for each PDGFRβ protomer.
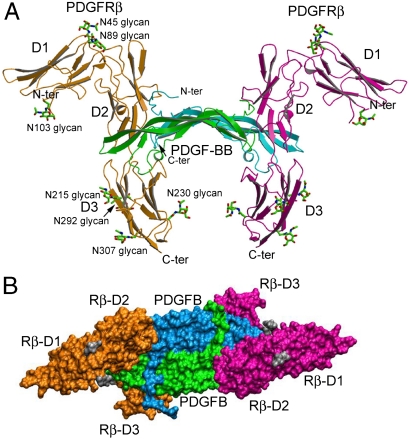
The 2:2 PDGF-B/PDGFRβ complex contains an antiparallel PDGF-B dimer symmetrically flanked by two monomeric PDGFRβ molecules (Fig. 2). The core scaffold of the PDGF-B dimer is unchanged from the free PDGF-B structure (PDB code 1PDG) (17), and is also similar to the PDGF-A dimer in the PDGF-A/propeptide complex. At the ends of the PDGF-B dimer, similar to PDGF-A, two clamps are formed by three interstrand loops (L1 for residues 25–38 and L3 for residues 78–81 of the protruding protomer, and L2 for residues 53–58 of the receding protomer) and the C-terminal segments. Each clamp interacts with a PDGFRβ molecule, thus each receptor molecule interacts with both ligand molecules.
Fig. 2.
Structure of the PDGF-B:PDGFRβ complex. (A) Side view of the ribbon model of the complex, with the PDGF-B protomers in green and blue, and the PDGFRβ protomers in pink and orange. The N-linked glycans attached to PDGFRβ are depicted as sticks. (B) Top view of the PDGF-B:PDGFRβ complex in surface representation.
PDGFRβ-D1-D3 is a string of three I-set Ig-like domains (D1, D2, and D3), which is bent at the D1-D2 boundary and is roughly straight at the D2-D3 boundary. Ligand binding is limited to the D2 and D3 domains. D1 is a small domain with short strands and, in the structure, contains three N-linked glycans. The FG loop of D1 is disordered. D2 is a distorted I-set Ig domain. No glycan is attached to D2. D3 is a canonical Ig domain containing four N-linked glycans. The D1-D2 boundary is supported by mostly hydrophobic interactions between D1 and D2, burying 1,060 Å2 of accessible surface area. The bent conformation between D1 and D2 is similar among PDGFRβ and KIT or FMS (Fig. S3). In comparison to the extensive D1-D2 interaction, there is only marginal contact between D2 and D3, which presents a large cleft at the D2-D3 boundary for interaction with PDGF-B.
The PDGF-B:PDGFRβ complex , which is dimerized by a cystine-knot-β-sheet fold ligand, has both similarities to and major differences from the other class III RTK complexes which are dimerized by four helix-bundle-fold ligands (Fig. S3A). The orientations of the D3 domains are similar in the SCF/KIT complex, the M-CSF/FMS complex, and the PDGF-B/PDGFRβ complex, however the orientations of D1 domains are dramatically different. Because D1-D2 is a rigid structural module, the distinct positions of D1 must originate from the different orientations of D2, which are positioned differently by the interaction with the ligands (Fig. S3 B and C).
The propeptide sequence of PDGF-B was included in the expression construct, but was not observed in the structure of the PDGF-B/PDGFRβ complex. Therefore, receptor binding apparently enabled or accelerated the extracellular cleavage and the displacement of propeptide from mature PDGF-B (Fig. S1C). The absence of propeptide suggests that propeptide binding and receptor binding are mutually exclusive for PDGFs. Indeed, superposition of the PDGF-A/propeptide complex and the PDGF-B/PDGFRβ complex by overlaying the PDGFs revealed that the propeptide and the receptor occupy overlapping sites on PDGFs (Fig. S3D), both centered at the same large cluster of hydrophobic residues at the L3 loop and the base of the L1 loop (Fig. 1B and Fig. 3). It is likely that the large hydrophobic surface of PDGFs required for receptor binding, when exposed in the ER, is unfavorable for folding. The propeptides were likely evolutionarily designed to cover such surfaces, overcoming problems during secretion.
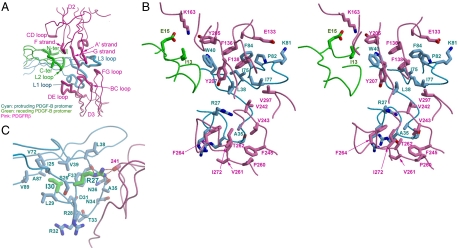
Fig. 3.
The PDGF-B:PDGFRβ interface. (A) An overview of the composite interface. The structural segments involved in binding are shown as thicker coils than the nonbinding segments, with the protruding PDGF-B protomer in cyan, the receding PDGF-B protomer in green, and PDGFRβ in pink. (B) Stereo view of the receptor-ligand interface in the same orientation, with main chains shown as coils and side chains shown as sticks. Note that upper patch and the lower patch of the interface are related by ∼110°. (C) The role of PDGF-B residues Arg27 and Ile30 (highlighted in green) in maintaining the receptor-binding conformation of the L1 loop. The view is rotated ∼70° from B.
The PDGF-B:PDGFRβ Interface.
Each PDGF-B:PDGFRβ interface buries 2,870 Å2 solvent-accessible surface area. On the PDGF-B side, the majority of the interface (1,950 Å2, or 67%) is contributed by the longer arm of the clamp, i.e., the L1 loop and L3 loop of the protruding protomer, whereas a smaller fraction (920 Å2, or 33%) is contributed by the shorter arm of the clamp, i.e., the L1 loop and the C terminus of the receding protomer (Fig. 3A). The N terminus of the receding PDGF-B protomer also makes marginal contact with the receptor. On the PDGFRβ side, the interface can be almost equally divided between the D2 domain and the D3 domain. The D2 domain contributes its lower A’GF β-sheet face to the upper side of the PDGF-B clamp, and also its CD loop to interact with the PDGF-B N terminus at the far edge of the interface. The PDGFRβ D3 domain contacts the lower side of the PDGF-B clamp with its BC, DE, and FG loops.
The large PDGF-B/PDGFRβ interface is a continuous and integral site, although it can be described as two adjacent, relatively flat subinterfaces related by ∼110°. The two subinterfaces are roughly divided at the midline of the PDGF-B clamp (Fig. 3). One subinterface, the upper patch, is formed mainly by the lower A’GF β-sheet of PDGFRβ-D2 docking onto the PDGF-B dimeric seam. The central feature of this subinterface is a large, hydrophobic, and aromatic-rich cluster, formed by the side chains of Tyr205, Tyr207, Phe136, Phe138 from PDGFRβ-D2 and Trp40, Leu38, Ile75, Ile77, Pro82, Phe84 from the protruding PDGF-B protomer (Fig. 3B, upper right). The receding protomer plays an auxiliary role at this subinterface by forming hydrophilic interactions with PDGFRβ (Fig. 3 and Table S3). Notably, Glu15 in the N-terminal segment of PDGF-B forms a salt bridge with Lys163, and likely charge attraction with Lys164, of PDGFRβ at the edge of the interface (Fig. 3B, upper left).
The second subinterface, the lower patch, is formed by the interactions between the membrane-distal loops of PDGFRβ-D3 and PDGF-B (Fig. 3 A and B). The PDGF-B interactions are mostly contributed by the L1 loop of the protruding PDGF-B protomer (Fig. 3B, lower part) (Table S3). The L1 loop of PDGF-B extends far from the cystine-knot β-sheet core, reaching to the waistline of the PDGFRβ-D3 β-sandwich. The interactions at the lower patch, like the upper patch, include a hydrophobic core, surrounded by hydrogen bonds and a salt bridge at the edge (Table S3). The continuous hydrophobic area of PDGFRβ-D3 is formed by Phe264, Ile272, Phe245, Val261, Pro260, Val243, and the Cγ atom of Thr262. Although the L1 loop of PDGF-B is highly hydrophilic, it manages to form a hydrophobic area by positioning the hydrophilic sidechains radially around Ala35, so that the aliphatic parts of these side chains, including Asn34, Asn36, Thr33, Arg28, and Arg32, form a flat hydrophobic surface facing the large hydrophobic area on PDGFRβ-D3. While most hydrophilic interactions are located at the edge of the interface, there is one notable exception: the terminal guanidino group of Arg27 is buried at the interface, forming hydrogen bonds with the main chain oxygen atom of PDGFRβ Glu241.
The PDGF-B/PDGFRβ interface can be used to reconcile a wide array of previous mutagenesis and deletion mapping data on PDGF-PDGFR interactions (18–22), which identified the L1, L2, and L3 loops as part of the receptor-binding epitope. In particular, Clements et al. mutated throughout the PDGF-B surface, and found that most mutations have only minor effects on receptor binding, but that Arg27 and Ile30 are two major determinants for PDGF-B:PDGFRβ interaction (19). In the complex, Arg27, as discussed above, forms hydrogen bonds with the main chain oxygen atom of PDGFRβ Glu241. But more importantly, the guanidino group of Arg27 also forms hydrogen bonds with two main chain oxygen atoms from PDGF-B itself (Fig. 3C). These hydrogen bonds (Arg27 Nϵ—Phe37 O, and Arg27 Nη2—Ala35 O) are crucial and serve to lock the conformation of the L1 loop, enabling the formation of the hydrophobic patch centered at Ala35 for receptor contact. In a similar fashion, although Ile30 does not have direct contact with the receptor, its side chain inserts into the large hydrophobic pocket formed by PDGFB Phe37, Val39, Val72, Val89, Ala87, and Val44, allowing the formation of the kinked conformation at the beginning of the L1 loop, and preparing it for receptor contact (Fig. 3C). Taken together, these observations indicate that the PDGF-B L1 loop conformation, rather than the point-to-point contacts at the interface, is the structural determinant for PDGF-B:PDGFRβ binding.
The Specificity and Promiscuity of PDGF-PDGFR Interaction.
PDGFRα can be activated by homodimeric PDGF-AA, PDGF-BB, and PDGF-CC, and heterodimeric PDGF-AB. PDGFRβ can be activated by PDGF-BB and PDGF-DD (reviewed in ref. 15). This interaction pattern indicates that PDGFRα is more promiscuous than PDGFRβ, and PDGF-B is more promiscuous than PDGF-A. The sequence identity between PDGFRα and PDGFRβ is only 30%. Nevertheless, most of the hydrophobic residues involved in PDGF contact are conservative substitutions (Fig. S4). Notably, among the large number of aromatic residues in the PDGFRβ ligand-binding surface, all but one (Tyr207) are substituted to smaller residues (Tyr205Asn, Phe136Leu, Phe138Ile, Tyr270Ile, Phe245Leu, and Phe264Glu) in PDGFRα. The aromatic side chains are bulkier and more difficult to rotate, imposing more stringent requirements on the ligands. This imposed stringency may explain why PDGFRβ is more selective than PDGFRα in PDGF binding.
The hydrophobic residues for contacting receptors are roughly similar between PDGF-A and PDGF-B (Fig. S4). The major difference between PDGF-A and PDGF-B is at the edge of the receptor-binding surface, where a large number of long-chain hydrophilic residues in PDGF-B are substituted by smaller residues in PDGF-A (Glu15Val, Arg28Ser, Arg32Pro, Asn34Ser, Asn55Thr, Arg56Ser, Asn57Ser, Arg73Ala, and Lys98Ala, PDGF-B numbering) (Fig. S4). The opposite trend is not observed. This smaller residue substitution indicates a reduction of the number of possible hydrogen bonds and salt bridges for PDGF-A. To compensate for such a reduction, the hydrophobic contact for the PDGF-A: receptor complex may require more complementarity, dictating why PDGF-A is more selective than PDGF-B in receptor binding.
The PDGF-B Conformational Change upon Receptor Binding.
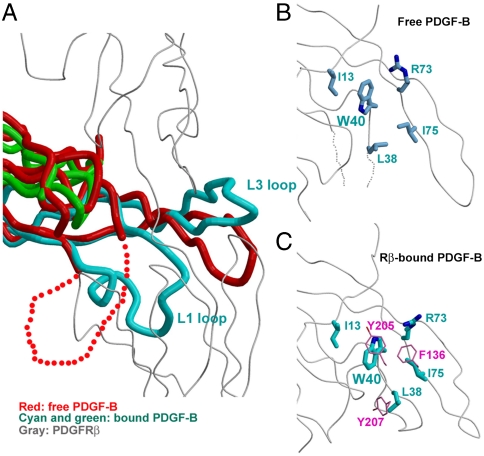
While the central β-strands of the receptor-bound PDGF-B are unchanged from the free PDGF-B, there are significant conformational differences at the loops and the N-terminus between the two forms (Fig. 4A). The largest change is the structural formation of the L1 loop, the conformation of which is supported by interactions on two sides: intermolecularly on one side by the contact with PDGFRβ-D3 (Fig. 3B, lower part), and intramolecularly on the other side by the anchoring of the Leu29-Ile30 side chains into the PDGF-B hydrophobic pocket formed by Phe37-Val39-Val72-Val89-Ala87-Val44 (Fig. 3C). The hydrophobic pocket is well maintained in free PDGF-B already, so anchoring at this pocket is insufficient for maintaining the L1 conformation in the absence of receptor. The loop itself is exquisitely configured in the complex, with the aromatic ring of Phe37 at the center of the hydrophobic core, and numerous intraloop hydrogen bonds to maintain main chain peptide conformation (Fig. 3C). Therefore, this loop has an intrinsic propensity to fold into the observed conformation. In support of this notion, the L1 loop of the propeptide-bound PDGF-A, which is only marginally stabilized at its base by the interaction with the propeptide, is ordered and adopts a similar conformation to PDGF-B (Fig. S3D). Hence, the free PDGF-B L1 loop, which was disordered in that structure, may exist in equilibrium between folded and unfolded states, the former only selected by receptor binding.
Fig. 4.
The PDGF-B conformational change upon PDGFRβ binding. (A) Comparison between the free PDGF-B (PDB code 1PDG) dimer (red) and the PDGFRβ-bound PDGF-B dimer (cyan for the protruding protomer and green for the receding protomer) shows that PDGFRβ-binding induces the structural organization of the previously disordered large L1 loop, and a rotation of the protruding L3 loop. (B) and (C) Comparison of PDGF-B Trp40 and its neighboring residues in the free form and receptor-bound form shows a 180° flipping of Trp40, as induced by its interactions with PDGFRβ Tyr205, Tyr207 and Phe136 (pink).
Another notable conformational change in PDGF-B is the flipping of the large aromatic side chain of Trp40 (Fig. 4 B and C). This side chain contacts Tyr205, Tyr207, and Phe136 of PDGFRβ. But in the conformation observed in the free PDGF-B structure, it would clash with the incoming PDGFRβ Tyr205. The possibility of PDGF-B Trp40 to adopt multiple conformations can increase the plasticity of the PDGF-B receptor-binding surface, a factor probably important in its ability to bind both PDGFRα and PDGFRβ. Similarly, the L3 loop of PDGF-B which harbors the Ile75-Ile77-Pro82-Phe84 hydrophobic cluster can rearrange easily, swinging as a unit toward the incoming receptor, forming interactions with PDGFRβ Phe138.
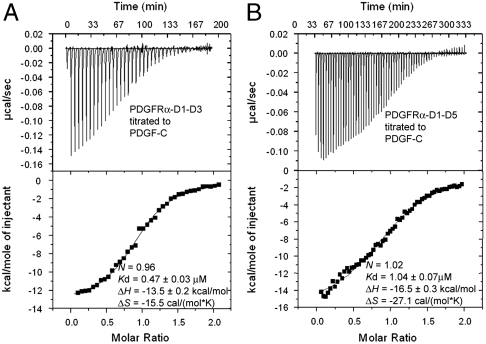
The structural organization of the L1 loop, and the conformational plasticity of the L3 loop and the hydrophobic surfaces, would be expected to impose a significant entropic penalty on the PDGF-B: PDGFRβ binding, similar to the entropic penalty observed for the folding transition of some proteins upon DNA binding (23). Because PDGF:PDGFR interactions are built on a common scaffold, most of these structural features should be shared by all PDGF:PDGFR interactions. Indeed, when we examined the binding between PDGF-C and PDGFRα using isothermal titration calorimetry (Fig. 5), we found a large entropic decrease in this reaction, probably indicative of PDGF-C becoming more ordered. The exact conformational change of PDGF-C, however, still awaits characterization.
Fig. 5.
Comparison of thermodynamic profiles of (A) PDGF-C:PDGFRα-D1-D3 binding and (B) PDGF-C:PDGFRα-D1-D5 binding, showing a negative contribution from PDGFRα-D4-D5.
The Role of PDGFR Membrane-Proximal Domains in the Signaling Assembly.
The class III RTKs have a five-domain extracellular configuration (6). While the first three domains of these receptors contain all the ligand-binding epitopes, the membrane-proximal D4 and D5 domains can play different roles in the formation of signaling assemblies and receptor activation. The D4-D5 domains of KIT, driven by the SCF/KIT interaction, undergo a lateral swing to result in a homotypic interaction to the equivalent domains in the other KIT receptor in the dimerized SCF:KIT complex (9). The FMS D4-D5, instead, using the homotypic interaction as a driving force, collaborates with the M-CSF/FMS interface to dimerize the two receptors (7). It has been suggested that the PDGFRβ D4-D5 also interact homotypically (24), but the contribution of these domains to ligand binding and receptor dimerization is unclear.
We compared the binding of PDGF-C, the only PDGF that could be expressed without propeptide, to PDGFRα, in the presence vs. absence of PDGFRα D4-D5 domains (Fig. 5). We were surprised to find that PDGF-C binding to the full extracellular segment was weaker than binding to only the first three domains of PDGFRα (1.04 μM for PDGFC:PDGFRα-D1-D5 and 0.47 μM for PDGFC:PDGFRα-D1-D3). The geometry of the PDGF-B:PDGFRβ-D1-D3 complex indicates that PDGFs are unlikely to reach receptor D4-D5 domains. Therefore the reduced PDGF-C binding in the presence of D4-D5 domains is due to receptor-receptor interactions. This homotypic interaction is likely similar to those of KIT D4-D5 and VEGFR2-D7, which have been elucidated crystallographically (9, 25); it should similarly be important for maintaining the geometry of the ligand/receptor complex, as have been shown for VEGF/VEGFR by mutagenesis and electron microscopy studies (26). However, the PDGFRα D4-D5 homotypic interaction is forced by ligand-receptor-binding, and may be weaker than the VEGFR2 D7 homotypic interaction which, despite burying a small interface, can be observed alone in the crystal (25). It has been reported that the PDGF-induced activation of PDGFRβ is compromised when Arg385 and Glu390, a pair of salt bridge-forming residues in D4, were mutated to alanine (24). In PDGFRα, the glutamate residue is substituted by aspartate. The formation of the corresponding salt bridge in PDGFRα should require the two D4 domains to move closer together than in PDGFRβ, as well as compared to KIT and FMS, which have glutamate at this position. Because of the lack of a highly complementary interface between D4 domains, as exemplified by the SCF/KIT complex (9), this close proximity between PDGFRα D4 domains can be energetically unfavorable, despite the fact that this geometry is required for kinase activation.
Implications for VEGFR/PDGFR Signaling and VEGF/PDGF Antagonism.
VEGF/PDGF signaling is conserved in invertebrates and vertebrates (3), and the N-terminal Ig domains of PDGFRs/VEGFRs implicated in ligand binding are likely to be structurally conserved. The PDGF-B/PDGFRβ structure indicates that the D1 domains of PDGFR/VEGFRs are generally not involved in ligand binding, but serve as a cap for the ligand-binding D2 domains. The orientation of D2 and D3 domains in the complex should be similar for all VEGFRs and PDGFRs, as supported by the structural comparison between the PDGF-B/PDGFRβ complex and the recently determined VEGF-C/VEGFR2 complex (11) (Fig. S3A and Fig. S5). However, VEGFR2-D2 is more tilted and shifted outward from the center of the complex than PDGFRβ-D2 and VEGFR2-D3 is more shifted to the side than PDGFRβ-D3. These differences are coupled to the structural variations of the ligands, such as at the N-terminal segments (helical in VEGFs vs. extended in PDGFs), the L1 loop (the L1 loops of VEGFs are shorter than of PDGFs), and the L3 loop (the L3 loops of VEGFs are swung down but that of PDGF-B is straight relative to the central β-sheets) (Fig. S5). Structural comparison also revealed that the composition of the receptor/ligand interface is vastly different between PDGF-B/PDGFRβ and VEGF-C/VEGFR2 (11). The difference is, however, consistent with the low sequence similarity and the conformational variability of their interface-participating segments (Fig. S4 and Fig. S5). Therefore, the VEGF/VEGFR and PDGF/PDGFR pairs may have diverged significantly in evolution for separate functions. A previous report indicated that VEGF could signal through PDGFR in bone-marrow-derived mesenchymal cells (27). However, we were unable to detect binding between recombinant VEGF and PDGFRs biochemically.
The PDGF-B/PDGFRβ complex is useful for guiding the design of receptor decoys for VEGF/PDGF inhibition. For instance, the currently used VEGF-trap is a combination of the VEGFR1-D2 and VEGFR2-D3 domains (28). Because there is a hydrophobic interface between D1 and D2, the D2–D3 decoy may have an exposed hydrophobic surface due to the absence of D1. Unnecessary hydrophobic surfaces on the protein can result in nonspecific adhesion and the deposition at the drug administration site, compromising the drug’s phamacokinetic profile. In PDGFRβ, the hydrophobic D1–D2 interaction is contributed mostly by the D1 residues Tyr117, Phe119 and the D2 residues Leu128, Thr152, and Pro154, which are conserved in PDGFRs/VEGFRs. These D2 residues on the VEGF-trap could be replaced by charged or more polar residues to improve behavior in solution. The PDGF-B: PDGFRβ structure also suggests a possibility of designing higher-affinity, broader-spectrum PDGF inhibitors by combining PDGFRα and PDGFRβ structural features, especially by replacing the aromatic residues at the interface with smaller aliphatic residues. In addition, the D2–D3 junction of PDGFRs, with little direct interaction between D2 and D3, appears to be prone to hinge flexibility when the ligand is absent. This flexibility may be unfavorable for ligand binding due to entropy. Engineering a structurally rigid D2–D3 junction, perhaps corresponding closely to its conformation in the ligand-bound state, may render the binding of the receptor decoy to ligands more entropically favorable. Lastly, the conserved prosequences of PDGFs preceding the growth factor domains are competent in ligand binding, and are simpler than antibodies or receptor decoys. Engineering a high-affinity, prosequence-mimicking PDGF-trap can also be envisioned to target PDGF signaling.
Methods
Recombinant Protein Expression and Crystallization.
The proteins were expressed either from baculovirus-infected insect cells, or from mammalian cells using the BacMam method previously described (14). All proteins were glycan-minimized before crystallization. Details are described in SI Text.
Data Collection and Structure Determination.
The crystallographic data were measured and processed and the structure was determined as described in SI Text. Crystallographic data statistics are listed in Table S1.
Isothermal Titration Calorimetry.
Calorimetric titrations were carried out on a VP-ITC calorimeter (MicroCal) at 30 °C as described in SI Text. The data were processed with the MicroCal Origin 5.0 software.
Supplementary Material
Acknowledgments.
We thank K.C. Garcia for kindly providing the BacMam reagents and guidance. X.H. is supported by the Brain Tumor Society and the National Institutes of Health (NIH) Grant 1R01GM078055. The Structural Biology Facility is supported by the R.H. Lurie Comprehensive Cancer Center of Northwestern University. Data were measured at the LS-CAT beamline 21-ID-D at the Advanced Photon Source (APS), Argonne, IL.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3MJK and 3MJG).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000806107/-/DCSupplemental.
References
- 1.Kohler N, Lipton A. Platelets as a source of fibroblast growth-promoting activity. Exp Cell Res. 1974;87:297–301. doi: 10.1016/0014-4827(74)90484-4. [DOI] [PubMed] [Google Scholar]
- 2.Ross R, Glomset J, Kariya B, Harker L. A platelet-dependent serum factor that stimulates the proliferation of arterial smooth muscle cells in vitro. Proc Natl Acad Sci USA. 1974;71:1207–1210. doi: 10.1073/pnas.71.4.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ostman A. PDGF receptors-mediators of autocrine tumor growth and regulators of tumor vasculature and stroma. Cytokine Growth F R. 2004;15:275–286. doi: 10.1016/j.cytogfr.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Fredriksson L, Li H, Eriksson U. The PDGF family: four gene products form five dimeric isoforms. Cytokine Growth F R. 2004;15:197–204. doi: 10.1016/j.cytogfr.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Liu H, Focia PJ, Shim AH, He X. Structure of macrophage colony stimulating factor bound to FMS: diverse signaling assemblies of class III receptor tyrosine kinases. Proc Natl Acad Sci USA. 2008;105:18267–18272. doi: 10.1073/pnas.0807762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H, Chen X, Focia PJ, He X. Structural basis for stem cell factor-KIT signaling and activation of class III receptor tyrosine kinases. EMBO J. 2007;26:891–901. doi: 10.1038/sj.emboj.7601545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuzawa S, et al. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130:323–334. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]
- 10.McDonald NQ, Hendrickson WA. A structural superfamily of growth factors containing a cystine knot motif. Cell. 1993;73:421–424. doi: 10.1016/0092-8674(93)90127-c. [DOI] [PubMed] [Google Scholar]
- 11.Leppanen VM, et al. Structural determinants of growth factor binding and specificity by VEGF receptor 2. Proc Natl Acad Sci USA. 2010;107:2425–2430. doi: 10.1073/pnas.0914318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergers G, Song S, Meyer-Morse N, Bergsland E, Hanahan D. Benefits of targeting both pericytes and endothelial cells in the tumor vasculature with kinase inhibitors. J Clin Invest. 2003;111:1287–1295. doi: 10.1172/JCI17929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erber R, et al. Combined inhibition of VEGF and PDGF signaling enforces tumor vessel regression by interfering with pericyte-mediated endothelial cell survival mechanisms. FASEB J. 2004;18:338–340. doi: 10.1096/fj.03-0271fje. [DOI] [PubMed] [Google Scholar]
- 14.Dukkipati A, Park HH, Waghray D, Fischer S, Garcia KC. BacMam system for high-level expression of recombinant soluble and membrane glycoproteins for structural studies. Protein Expres Purif. 2008;62:160–170. doi: 10.1016/j.pep.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heldin CH, Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev. 1999;79:1283–1316. doi: 10.1152/physrev.1999.79.4.1283. [DOI] [PubMed] [Google Scholar]
- 16.Reigstad LJ, Varhaug JE, Lillehaug JR. Structural and functional specificities of PDGF-C and PDGF-D, the novel members of the platelet-derived growth factors family. FEBS J. 2005;272:5723–5741. doi: 10.1111/j.1742-4658.2005.04989.x. [DOI] [PubMed] [Google Scholar]
- 17.Oefner C, D’Arcy A, Winkler FK, Eggimann B, Hosang M. Crystal structure of human platelet-derived growth factor BB. EMBO J. 1992;11:3921–3926. doi: 10.1002/j.1460-2075.1992.tb05485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersson M, et al. Involvement of loop 2 of platelet-derived growth factor-AA and -BB in receptor binding. Growth Factors. 1995;12:159–164. doi: 10.3109/08977199509028962. [DOI] [PubMed] [Google Scholar]
- 19.Clements JM, et al. Two PDGF-B chain residues, arginine 27 and isoleucine 30, mediate receptor binding and activation. EMBO J. 1991;10:4113–4120. doi: 10.1002/j.1460-2075.1991.tb04988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaRochelle WJ, Pierce JH, May-Siroff M, Giese N, Aaronson SA. Five PDGF B amino acid substitutions convert PDGF A to a PDGF B-like transforming molecule. J Biol Chem. 1992;267:17074–17077. [PubMed] [Google Scholar]
- 21.Mahadevan D, et al. Structural role of extracellular domain 1 of alpha-platelet-derived growth factor (PDGF) receptor for PDGF-AA and PDGF-BB binding. J Biol Chem. 1995;270:27595–27600. doi: 10.1074/jbc.270.46.27595. [DOI] [PubMed] [Google Scholar]
- 22.Ostman A, Andersson M, Backstrom G, Heldin CH. Assignment of intrachain disulfide bonds in platelet-derived growth factor B-chain. J Biol Chem. 1993;268:13372–13377. [PubMed] [Google Scholar]
- 23.Sieber M, Allemann RK. Thermodynamics of DNA binding of MM17, a “single chain dimer”of transcription factor MASH-1. Nucleic Acids Res. 2000;28:2122–2127. doi: 10.1093/nar/28.10.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, Yuzawa S, Schlessinger J. Contacts between membrane proximal regions of the PDGF receptor ectodomain are required for receptor activation but not for receptor dimerization. Proc Natl Acad Sci USA. 2008;105:7681–7686. doi: 10.1073/pnas.0802896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y, Xie P, Opatowsky Y, Schlessinger J. Direct contacts between extracellular membrane-proximal domains are required for VEGF receptor activation and cell signaling. Proc Natl Acad Sci USA. 2010;107:1906–1911. doi: 10.1073/pnas.0914052107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruch C, Skiniotis G, Steinmetz MO, Walz T, Ballmer-Hofer K. Structure of a VEGF-VEGF receptor complex determined by electron microscopy. Nat Struct Mol Biol. 2007;14:249–250. doi: 10.1038/nsmb1202. [DOI] [PubMed] [Google Scholar]
- 27.Ball SG, Shuttleworth CA, Kielty CM. Vascular endothelial growth factor can signal through platelet-derived growth factor receptors. J Cell Biol. 2007;177:489–500. doi: 10.1083/jcb.200608093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holash J, et al. VEGF-Trap: a VEGF blocker with potent antitumor effects. Proc Natl Acad Sci USA. 2002;99:11393–11398. doi: 10.1073/pnas.172398299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.