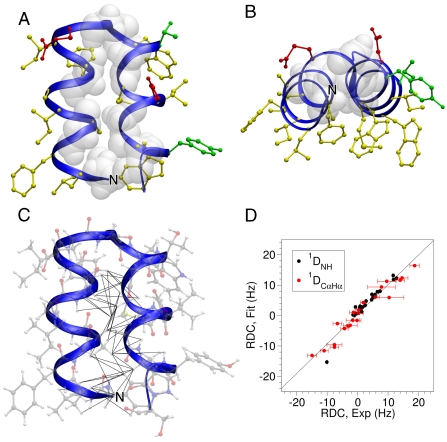
Fig. 2.
Structure of HAfp1–23. (A) Longitudinal view with the hydrophobic side chains drawn in yellow, polar side chains in green, acidic side chains in red, and Gly residues shown in white van der Waals representation. The interhelical angle between helix A (residues 2–12) and helix B (residues 14–22) is 158 °, and the interhelical distance calculated using the program interhlx (K. Yap, University of Toronto) is 6.3 Å. The amino terminus is marked N.(B) Lateral view. (C) Longitudinal view showing all atoms, and interhelical NOEs below 5 Å as black lines. (D) Singular value decomposition fit of the pH 4.0 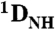 and
and 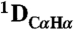 RDCs to the refined pH 7.4 structure. The
RDCs to the refined pH 7.4 structure. The 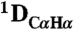 couplings have been scaled by -0.484 relative to
couplings have been scaled by -0.484 relative to 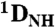 to account for the relative internuclear dipolar interaction strength. The Q-factor of the pH 4.0 data to the pH 7.4 structure is 22.3%.
to account for the relative internuclear dipolar interaction strength. The Q-factor of the pH 4.0 data to the pH 7.4 structure is 22.3%.