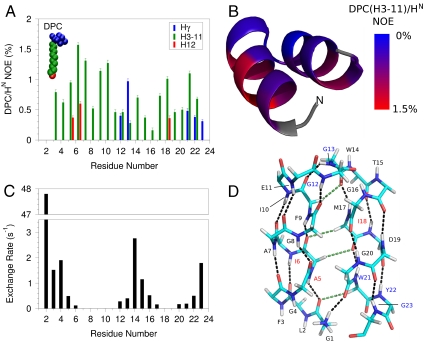
Fig. 3.
HAfp interactions with the micelle and with water at pH 7.4. (A) Strength of NOE interactions between perdeuterated, amide-protonated HAfp1–23 and DPC micelle protons. Amides with cross peaks to choline methyl protons (Hγ) are at the lipid-water interface. Residues with stronger intensity to the DPC methylene (H3-H11) and methyl (H12) protons are closer to the lipid core of the micelle. A NOE mixing time of 100 ms was used, and the plotted intensities have been scaled by the intensity of the diagonal amide resonance. DPC/HN NOEs at pH 7.4 and 4.0 are compared in Fig. S5C. (B) NOE intensity between the DPC H3-H11 methylene protons and the backbone HN protons of the fusion peptide mapped on the structure, with red corresponding to strongest NOEs. Residues in gray could not be detected due to fast exchange of the amide/amino protons with water. The amino terminus is marked N. (C) Solvent hydrogen exchange rates of the backbone amide protons. (D) Hydrogen bonding pattern in the HAfp1–23 structure. Residues Gly1 to Gly12 and Trp14 to Gly23 have well satisfied αhelical hydrogen bonds, marked by black dashed lines. CαH-O = C hydrogen bonds are shown as green dashed lines. Hydrogen bonding between the 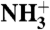 of the N-terminal Gly1 and the carbonyl oxygen of Gly20 is also suggested by the structure. Inclusion of the N-terminal hydrogen bond and the CαH-O = C hydrogen bonds does not adversely affect experimental statistics of the refined structure (SI Text). Blue residue labels indicate proximity to the DPC choline; red labels are for residues with NOEs to the DPC methyl groups.
of the N-terminal Gly1 and the carbonyl oxygen of Gly20 is also suggested by the structure. Inclusion of the N-terminal hydrogen bond and the CαH-O = C hydrogen bonds does not adversely affect experimental statistics of the refined structure (SI Text). Blue residue labels indicate proximity to the DPC choline; red labels are for residues with NOEs to the DPC methyl groups.