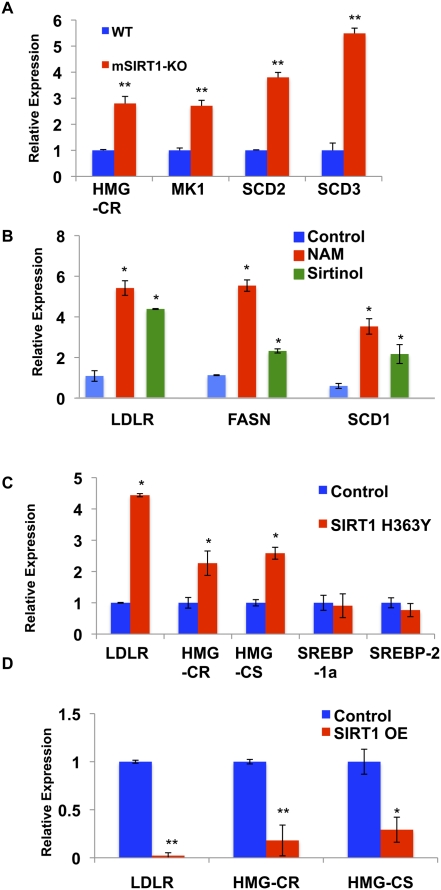
Figure 4.
Mammalian SIRT1 represses SREBP-1 and SREBP-2 target gene expression. (A) SREBP target genes are expressed at higher levels in SIRT1 knockout MEFs when compared with the wild-type cells, as judged by qRT–PCR; β-actin served as the control of total RNA. (MK1) Mevalonate kinase-1; (SCD2) stearoyl-CoA desaturase-2; (SCD3) stearoyl-CoA desaturase-3. (B) Treatment of human IMR-90 fibroblasts with the sirtuin inhibitors nicotinamide (NAM; 20 mM) or sirtinol (0.1 mM) for 5 h resulted in increased expression of SREBP target genes by qRT–PCR. Data were first normalized by β-actin, then normalized by the control (0.1% DMSO), and expressed as mean ± SD. (SCD1) Stearoyl-CoA desaturase-1. (C) Expression of a dominant-negative form of SIRT1 (H363Y) in HEK293T cells increased the transcription of the LDLR, HMG-CR, HMG-CS, and SCD1 genes as assessed by qRT–PCR. SREBP-1a or SREBP-2 transcriptional levels were unchanged. GAPDH served as a loading control of total RNA. (D) Overexpression of SIRT1 decreases expression of SREBP target genes in HEK293T cells. SIRT1 and SIRT1(H363Y) (Fig. 4C) were expressed at similar levels. Error bars represent standard deviations between cDNA made from independently treated cells. (*) P < 0.05; (**) P < 0.01.