Figure 1.
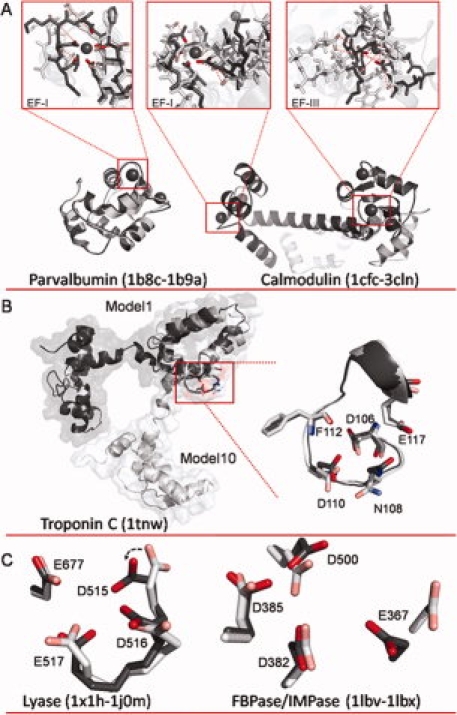
A: Overlays of apo and holo structures of parvalbumin and calmodulin (CaM). For parvalbumin (apo 1B8C.pdb and holo 1B9A.pdb), alignment indicates little change in the backbone conformation, but more significant restructuring in the EF-I binding site where side chains rotate inward to form the binding pocket. Conversely, for CaM (apo 1CFC.pdb and holo 3CLN.pdb), significant restructuring is observed both globally and within the binding sites. CaM sites EF-1 and EF-III were modeled individually by aligning only along the binding site residues in each loop for the apo and holo structures. Dashed red lines indicate distance between key binding ligands between the two structures. B: Model 1 (dark gray) and Model 10 (light gray) of NMR structure of troponin C (1TNW.pdb) aligned along residues 106–117. C: Overlay of calcium binding pocket of lyase (left) in apo (1X1H.pdb, light gray) and holo (1J0M.pdb, dark gray) structures, and overlay of calcium binding pocket of FBPase/IMPase (right) in apo (1LBV.pdb, light gray) and holo (1LBX.pdb, dark gray) structures. An interactive view is available in the electronic version of the article. PRO394 Figure 1
