Figure 3.
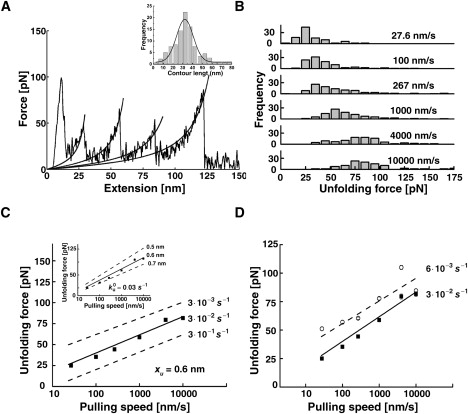
Forced unfolding of poly-AcP. (A) A typical force-extension curve obtained by stretching individual AcP polymers at 100 nm/s. The high force peak seen at the beginning of the extension profile reflects nonspecific interactions between the AFM tip and the mounting surface. The solid lines superimposed on the rising parts of the peaks are fits to a worm-like-chain model. (A, inset) Contour length increments upon domain unraveling obtained from the fitting (vc = 267 nm/s). (B) Frequency histograms of unfolding forces recorded at different pulling speeds. (C) Dependence of the most probable force for unfolding, taken as the maximum of the unfolding force distributions, on the pulling speed. The best fit to the data from the Monte Carlo simulations (solid lines in the main figure and inset) was obtained using = 0.03 s−1 (main figure) and xu = 0.6 nm (inset). It was previously shown that very high pulling speeds could be associated with distance-dependent drag forces, which may lead to underestimation of the unfolding force at such speeds (67,68). Our analysis reveals that the deviation expected, even for the highest pulling speed used in the experiments described in this work, lies within the thermal noise error. (D) Force spectra obtained for poly-AcP in the absence (solid rectangles, solid line) or presence (open circles, dashed line) of 10 mM Pi. The presence of the ligand stabilizes the native structure of the protein, leading to deceleration of the unfolding reaction. However, the position of the transition state ensemble along the force-set unfolding pathways is not affected by the ligand, as evidenced by the fact that the slope of the force spectrum is unaltered. Albeit not seen in all data points shown, SE bars are included (each data point represents the average of ∼70–250 data points cumulatively acquired in two to three independent experiments).
