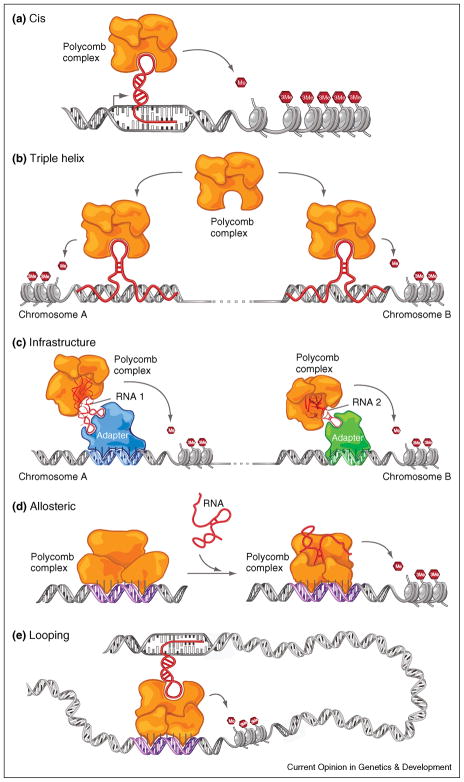
Figure 2.
Models how large non-coding RNAs could impart specificity to chromatin modifications (a) Cis model: at the site of RNA transcription, chromatin-modifying complexes could be recruited directly or indirectly and exert their function locally in cis. (b) Triple helix model: Formation of a RNA-DNA triplex could serve as a beacon to recruit directly or indirectly epigenetic modifying complexes to specific sites. (c) Infrastructure model: large non-coding RNAs could serve as an architectural scaffold that directly or indirectly links chromatin complexes to DNA binding proteins or transcription factors. (d) Allosteric model: RNA could bind directly or indirectly to chromatin remodeling proteins that change the shape of the protein or protein complex, activating/inactivating the complex and confer site specificity. (e) Looping model: RNA could serve as a scaffold to establish long range interactions via DNA looping and recruiting chromatin modifying complexes to distal sites of action in cis and or inter-chromosomal interactions (not shown).