Table 3. Cross-learning results.
| Kernel | AIMed | BioInfer | HPRD50 | IEPA | LLL | |||||||||||||||
| AUC | P | R | F | AUC | P | R | F | AUC | P | R | F | AUC | P | R | F | AUC | P | R | F | |
| SL | 77.5 | 28.3 | 86.6 | 42.6 | 74.9 | 62.8 | 36.5 | 46.2 | 78.0 | 56.9 | 68.7 | 62.2 | 75.6 | 71.0 | 52.5 | 60.4 | 79.5 | 79.0 | 57.3 | 66.4 |
| SpT | 56.8 | 20.3 | 48.4 | 28.6 | 64.2 | 38.9 | 48.0 | 43.0 | 60.4 | 44.7 | 77.3 | 56.6 | 54.2 | 41.6 | 19.6 | 15.5 | 50.5 | 48.2 | 83.5 | 61.2 |
| kBSPS | 72.1 | 28.6 | 68.0 | 40.3 | 73.3 | 62.2 | 38.5 | 47.6 | 78.3 | 61.7 | 74.2 | 67.4 | 81.0 | 72.8 | 68.7 | 70.7 | 86.8 | 83.7 | 75.0 | 79.1 |
| cosine | 65.4 | 27.5 | 59.1 | 37.6 | 61.3 | 42.1 | 32.2 | 36.5 | 71.2 | 63.0 | 56.4 | 59.6 | 57.0 | 46.3 | 31.6 | 37.6 | 66.9 | 80.3 | 37.2 | 50.8 |
| edit | 62.8 | 26.8 | 59.7 | 37.0 | 61.0 | 53.0 | 22.7 | 31.7 | 60.7 | 58.1 | 55.2 | 56.6 | 62.1 | 58.1 | 45.1 | 50.8 | 57.6 | 68.1 | 48.2 | 56.4 |
| APG | 77.6 | 30.5 | 77.5 | 43.8 | 69.6 | 58.1 | 29.4 | 39.1 | 84.0 | 64.2 | 76.1 | 69.7 | 82.4 | 78.5 | 48.1 | 59.6 | 86.5 | 86.4 | 62.2 | 72.3 |
| Fayruzov et al. | 72.0 | 40.0 | 70.0 | 31.0 | 75.0 | 56.0 | 68.0 | 29.0 | 74.0 | 39.0 | ||||||||||
Classifiers are trained on the ensemble of four corpora and tested on the fifth one. Rows correspond to test corpora. Best results are typeset in bold (differences under 1 base point are ignored). We show for reference the results with the combined full kernel of [25], taken from [38]. AUC, precision, recall, and F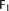 -score in percent.
-score in percent.
