Table I.
Structures, Inhibitory Potencies, and Simulation Data for Studied Inhibitors of MMP-3.
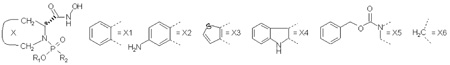 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No | X | R1 | R2 | log(1/Ki) | Linear Response terms in Steps 3 and 4 −Δ<x>, E in kcal/mol and SASA in Å2 | SD a | Bond length (Å) | Angle b | |||||||
| Exp. | Calc. | EvdW | Eel | EQM/MM. | SASA | O1-Zn | O2-Zn | O1-Zn-N2 | O1-Zn-N3 | N2-Zn-N3 | |||||
| 1 | X1 | CH2CH3 | 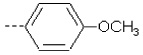 |
8.284 | 8.200 | 11.609 | 50.30 | 1890.75 | 464.46 | 0.155 | 1.844 | 2.017 | 104.6 | 167.5 | 87.9 |
| 2 | X1 | CH3 | 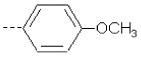 |
8.530 | 8.279 | 388.912 | 90.60 | 1924.28 | 477.20 | 0.765 | 1.956 | 1.947 | 104.9 | 166.3 | 88.8 |
| 3 | X1 | 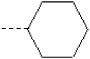 |
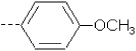 |
8.285 | 7.986 | 178.393 | 34.05 | 1863.53 | 428.70 | 0.456 | 1.946 | 1.948 | 94.8 | 179.0 | 84.3 |
| 4 | X1 | 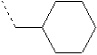 |
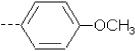 |
8.614 | 8.383 | 20.535 | 32.80 | 2067.29 | 491.82 | 0.213 | 1.965 | 1.953 | 108.0 | 164.1 | 86.9 |
| 5 | X1 | 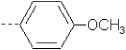 |
7.893 | 7.963 | 45.864 | 89.39 | 2493.80 | 411.07 | 0.318 | 1.947 | 1.947 | 100.6 | 171.4 | 87.8 | |
| 6 | X1 | (CH2)2N(CH2CH3)2 | 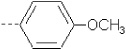 |
7.084 | 7.544 | 176.487 | 19.79 | 917.12 | 373.92 | 0.143 | 1.716 | 1.931 | 135.0 | 119.2 | 104.3 |
| 7 | X1 | 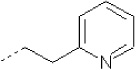 |
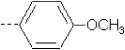 |
8.193 | 7.915 | 58.753 | 107.98 | 4776.15 | 353.47 | 0.202 | 1.849 | 2.031 | 102.1 | 164.8 | 91.2 |
| 8 | X1 | (CH2)2OCH2CH3 | 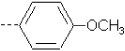 |
8.265 | 8.314 | 80.24 | 60.85 | 4873.99 | 419.29 | 0.322 | 1.947 | 1.943 | 103.9 | 161.6 | 91.1 |
| 9 | X1 | CH2CH3 | 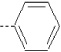 |
7.457 | 7.342 | 20.15 | 11.66 | 1720.62 | 322.14 | 0.26 | 1.946 | 1.946 | 106.6 | 167.2 | 86.6 |
| 10 | X1 | CH2CH3 | 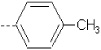 |
7.137 | 7.373 | 354.63 | 38.08 | 741.11 | 348.69 | 0.438 | 1.843 | 2.017 | 105.3 | 161.9 | 89.7 |
| 11 | X1 | CH2CH3 | 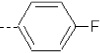 |
7.191 | 7.229 | 40.65 | 78.51 | 717.76 | 324.64 | 0.314 | 1.874 | 2.017 | 110.8 | 161.5 | 86.9 |
| 12 | X1 | CH2CH3 | 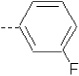 |
7.289 | 7.235 | 199.06 | 82.27 | 1341.75 | 312.27 | 0.743 | 1.867 | 1.987 | 115.2 | 153.8 | 90.3 |
| 13 | X1 | CH2CH3 | 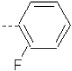 |
7.321 | 7.397 | 193.55 | 20.27 | 1659.06 | 332.94 | 0.395 | 1.875 | 1.99 | 104.5 | 167.8 | 85.7 |
| 14 | X1 | CH2CH3 | 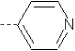 |
6.721 | 7.089 | 11.25 | 82.37 | 366.37 | 308.51 | 0.393 | 1.846 | 2.017 | 99.8 | 165 | 90.7 |
| 15 | X1 | CH2CH3 | 6.790 | 7.035 | 352.53 | 57.27 | 482.85 | 296.74 | 0.112 | 1.866 | 1.988 | 113.3 | 152.9 | 93.5 | |
| 16 | X1 | CH2CH3 | 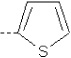 |
7.174 | 7.131 | 15.46 | 93.94 | 910.25 | 303.89 | 0.246 | 1.797 | 2.071 | 115.0 | 154.3 | 90.4 |
| 17 | X1 | CH2CH3 | 7.529 | 7.382 | 51.40 | 27.01 | 957.79 | 345.61 | 0.541 | 1.81 | 1.593 | 137.4 | 130.7 | 91.9 | |
| 18 | X1 | CH2CH3 | 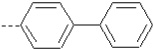 |
8.361 | 8.555 | 71.37 | 60.94 | 7570.98 | 401.89 | 0.281 | 1.875 | 1.995 | 119.3 | 150.4 | 89.7 |
| 19 | X1 | CH2CH3 | 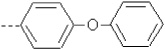 |
9.276 | 9.474 | 408.72 | 24.23 | 8665.43 | 534.54 | 0.101 | 1.849 | 2.026 | 124.0 | 142.8 | 93.2 |
| 20 | X1 | CH2CH3 | 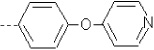 |
8.314 | 8.089 | 30.33 | 46.60 | 1497.50 | 454.18 | 0.131 | 1.758 | 2.039 | 116.0 | 148.8 | 93.5 |
| 21 | X1 | CH2CH3 | 9.301 | 9.432 | 36.51 | 67.38 | 7988.86 | 542.06 | 0.106 | 1.86 | 2.015 | 107.7 | 161.4 | 89.6 | |
| 22 | X1 | CH2CH3 | 7.936 | 7.545 | 46.63 | 135.80 | 3970.28 | 308.13 | 0.165 | 1.83 | 1.961 | 105.4 | 166.6 | 88.0 | |
| 23 | X2 | CH2CH3 | 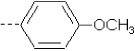 |
7.910 | 7.558 | 50.09 | 111.94 | 3283.05 | 325.14 | 0.17 | 1.864 | 1.988 | 94.7 | 163.6 | 84.1 |
| 24 | X3 | CH2CH3 | 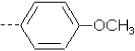 |
8.284 | 8.062 | 41.45 | 13.68 | 4667.35 | 380.83 | 1.78 | 2.052 | 2.028 | 104.8 | 158.2 | 85.5 |
| 25 | X4 | CH2CH3 | 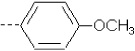 |
8.69 | 9.028 | 213.47 | 134.26 | 4963.11 | 538.76 | 0.399 | 1.821 | 1.875 | 125.2 | 130.7 | 101.1 |
| 26 | X5 | CH2CH3 | 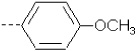 |
7.827 | 7.871 | 22.22 | 33.22 | 1625.96 | 414.26 | 0.213 | 1.861 | 2.057 | 110.4 | 161.0 | 86.0 |
| 27 | X6 | CH2CH3 | 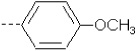 |
6.278 | 6.502 | 27.52 | 82.75 | 326.19 | 209.48 | 0.329 | 1.95 | 1.945 | 105.3 | 158.8 | 87.2 |
Standard deviation for the inhibitor atoms during the MD simulation.
Ideal values are 120° for trigonal bipyramid in all three cases, and 90°, 180°, and 90°, respectively, for the square pyramid. See Figure 1 for atom numbering.
