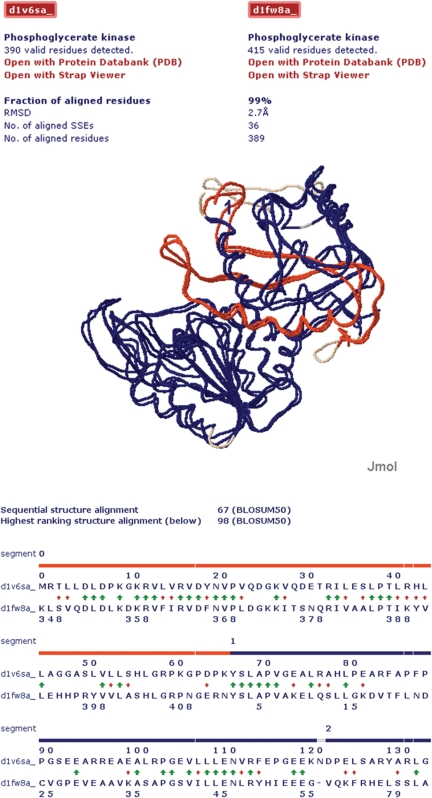
Figure 5.
Visualization of the circular permuted structure alignment result for the two proteins with PDB id 1V6S, chain A and 1FW8, chain A. Top part: protein structure alignment visualized with Jmol (see http://jmol.sourceforge.net for details). The aligned segments are shown in red and blue. Unaligned segments are shown in grey. Bottom part: illustration of the structure-based sequence alignment results evaluated with the BLOSUM50 substitution matrix (28). The color code is the same as in top part. The sequence break of the circular permutation (gap in blue band) has been detected between residues 66 and 67 in 1V6S, chain A. The green arrows indicate identical residue pairs and the red arrows residue pairs with a positive BLOSUM50 substitution score.