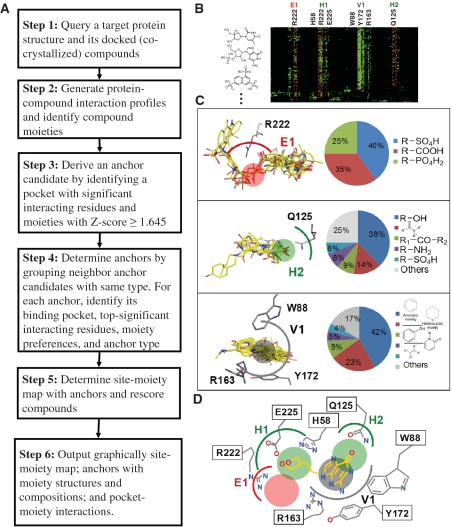
Figure 1.
Overview of the SiMMap server for the site-moiety map using herpes simplex virus type-1 thymidine kinase (TK) and 1000 docked compounds as the query. (A) Main procedure; (B) the merged protein–compound interaction profile; (C) the pocket–moiety interaction preferences of three anchors: E1 (electrostatic), H2 (hydrogen-bonding) and V1 (van der Waals). Each anchor consists of a binding pocket with conserved interacting residues, the moiety composition and anchor type; (D) the site-moiety map with four anchors.