Abstract
The enormous amount of data available in public gene expression repositories such as Gene Expression Omnibus (GEO) offers an inestimable resource to explore gene expression programs across several organisms and conditions. This information can be used to discover experiments that induce similar or opposite gene expression patterns to a given query, which in turn may lead to the discovery of new relationships among diseases, drugs or pathways, as well as the generation of new hypotheses. In this work, we present MARQ, a web-based application that allows researchers to compare a query set of genes, e.g. a set of over- and under-expressed genes, against a signature database built from GEO datasets for different organisms and platforms. MARQ offers an easy-to-use and integrated environment to mine GEO, in order to identify conditions that induce similar or opposite gene expression patterns to a given experimental condition. MARQ also includes additional functionalities for the exploration of the results, including a meta-analysis pipeline to find genes that are differentially expressed across different experiments. The application is freely available at http://marq.dacya.ucm.es.
INTRODUCTION
DNA microarrays have become an extensively used technique to analyze global gene expression profiles. Their widespread use quickly promoted the creation of gene expression data repositories such as the NCBI Gene Expression Omnibus (GEO; 1), which currently holds more than 10 000 experiments for over 500 organisms. In spite of this huge amount of information, in most cases, researchers still focus their analysis on individual experiments, making no use of such valuable resources. In this way, gene expression analyses are usually performed from a gene-centered perspective with the goal of identifying relevant sets of genes, e.g. genes differentially expressed between different conditions, which are then used to understand the biological mechanisms relevant in that experimental setup. However, besides this gene-to-phenotype approach, gene signatures can also be used to establish connections among different physiological conditions (2). For example, we can discover connections among diseases, drugs or pathways by similarities in their gene expression signatures; conditions inducing similar signatures might be modulating the same pathways, while conditions with opposite signatures might be involved in reverting the original phenotype.
During the last few years, previous works have proven the usefulness of mining large gene expression libraries to find similarities in gene expression signatures. Hughes et al. (3) showed that a compendium of gene expression data for diverse mutations and chemical treatments in yeast could be used for the functional annotation of small molecules and genes. Rhodes et al. (4) developed Oncomine, which integrates gene expression signatures from manually selected cancer microarray experiments from several sources. More recently, Lamb et al. (2) introduced the Connectivity Map, a searchable database that allows users to compare a query signature against a database populated with global signatures derived from human cell lines treated with different compounds. The Connectivity Map compares signatures using a statistic similar to the one in the Gene Set Enrichment Analysis (5), which is based on the ranks of the genes in the complete signature, as opposed to other applications such as L2L (6) or Exalt (7) that use threshold-based criteria. The rank statistic alleviates the high variability that comes from sub-setting the list of genes using a threshold. A more detailed comparison with related applications can be found in the additional material.
In this work, we present MARQ (Microarray Rank Query), a web-based application that allows researchers to query a signature database derived from GEO to find experiments that induce similar or opposite gene expression patterns to a given experiment. The signature database has been compiled from all GEO Datasets for five model organisms (Homo sapiens, Mus musculus, Rattus norvegicus, Saccharomyces cerevisiae and Arabidopsis thaliana). MARQ uses as input the standard output of most microarray studies, that is, a set of over-expressed and/or a set under-expressed genes, and uses a rank-based statistic to compare it with signatures in the database. MARQ also includes several functionalities that can be use for further exploration of results and to infer potential relationships among gene signatures in the database. The application is freely accessible at http://marq.dacya.ucm.es. The web portal includes tutorials of use and a detailed description of the web service interface and methods.
FEATURES AND FUNCTIONALITIES
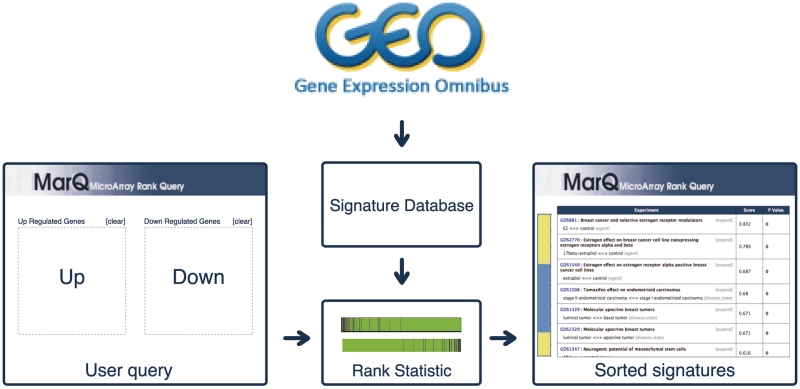
Figure 1 shows a general overview of the system. It has been organized in three main components: data input, data analysis and data visualization and exploration.
Figure 1.
MARQ is composed by three layers, the data input form (on the left-hand side) in which the user enters the query set of genes, the data analysis component that compares it against the database, and data visualization and exploration interface (on the right-hand side of the figure).
The input of the system consists of a query list of genes composed by a set of over-expressed and/or a set of under-expressed genes. This query is compared against all signatures in the database using a rank-based statistic and the output lists all signatures sorted by their similarity to the query. In this way, signatures with the highest positive scores represent experimental conditions that show similar over- and under-expression patterns to the query, while signatures with the highest negative scores are those that reverse the expression pattern of the query. The application also provides several features to explore and assess the significance of the results. Each signature is linked to a set of terms such as GO terms and words derived from its GEO description. These terms can be used to find datasets associated with the same terms and potentially sharing some biological characteristic, and to perform an enrichment analysis in order to determine which of these concepts are over-represented in the most significant signatures. These concepts may provide clues to understanding the molecular processes that are relevant in the query experiment.
Finally, MARQ also offers functionality for performing a rank-based differential expression analysis to detect genes that are commonly over or under-expressed across different signatures in the database.
Signature database construction
To build the gene signature database, we processed GEO Datasets for five different organisms: H. sapiens, M. musculus, R. norvegicus, S. cerevisiae and A. thaliana. The current version of MARQ contains 11 460 gene expression signatures from a total of 2050 GEO datasets and series from 355 platforms, including 708 datasets for human, 750 for mouse, 302 for yeast, 213 for rat and 77 for A. thaliana.
We retrieved from GEO all datasets associated to each organism using the NCBI E-utils and the GEOQuery package (8), Soft formatted gene expression data was loaded into the R environment, where the limma package was used to perform differential expression analysis among every pair of classes for each experimental factor. The gene signatures were then generated by sorting all genes in the array by their t-values, or fold-changes if the dataset did not have enough sample replicates to apply the statistical test. The direction of the comparison (A versus B or B versus A) was arbitrarily set, unless any of the classes were identified as a reference condition. Additionally, for two channel platforms we also performed one-class differential expression analysis, as each sample already represents a gene expression ratio.
For cross-platform comparisons, we used a common identifier to which all platform Ids were converted. Probes that could not be translated were removed from the analysis, and expression profiles were averaged for probes associated to the same gene. Using this framework, we generated a new set of gene-centered signatures.
Signature comparison
For a given query, we compute a similarity score for each signature in the database adapting the methodology reported in the Connectivity Map (2). This score reflects whether the up-regulated genes in the query tend to appear at the top of the database signature, while down-regulated genes tend to appear at the bottom (positive score) or vice versa (negative score). The query is composed of two sets of genes; the most highly over-expressed and/or the most highly under-expressed genes from a differential expression analysis. Signatures in the database represent the complete list of probes, from their corresponding dataset, sorted by differential expression analysis. A high positive score indicates that the database signature shares a significant proportion of over-expressed and under-expressed genes with the query, that is, ‘up genes’ in the query are also over-expressed in the signature and ‘down genes’ in the query are also under-expressed in the signature. Alternatively, a high negative score indicates that the signature has an antagonistic pattern, which means that ‘up genes’ in the query are under-expressed in the signature and ‘down genes’ in the query are over-expressed in the database signature. The score is calculated for the up and down-regulated genes using a weighted Kolmogorov–Smirnov-like statistic.
The significance of each score is determined using random permutations: the P-value associated with the score is the fraction of a large number of random lists of genes of the same length as the query that produce a larger score. Since thousands of signatures can be compared against each query a proportion of them could be false positives, so MARQ reports the P-values after making an FDR correction, as described in ref. (9). The randomization scheme used has been reported to be slightly flawed (10) in that it over-estimates significance, but it has been used successfully in similar settings, such as The Connectivity Map application (2). A complete description of the statistical tests and score computation are available at the application web site.
Connecting datasets through common biomedical concepts
In MARQ, each database signature is annotated with meta-information that allows us to connect them by common biological concepts, and to explore their relationships. These terms can be used for two purposes: to interactively find other signatures associated to the same biological concepts and explore their distribution in the sorted list, or to find which terms are over-represented among the most significant signatures.
Signatures are annotated with terms from different sources: terms contained in the description of experiments in GEO, and GO terms enriched in over-expressed and under-expressed genes in each signature. An enrichment score for each term associated to the significant signatures is calculated using the same rank-based statistical test as described above. The result of this analysis is a list of biological concepts with their corresponding P-values. Those terms with statistically significant P-values are representative of the most related signatures, and can provide information about the underlying biological processes, or highlight potential connections among different experimental conditions, for example, two different treatments that are activating the same pathway.
Comparing signatures to find common differentially expressed genes
One of the most powerful features of MARQ in exploring the results is a meta-analysis tool that can be used to find genes that are commonly deregulated across different database signatures. This analysis can be used, for example, to find out which genes that are over-expressed in a disease signature are under-expressed after a treatment signature. MARQ implements the Rank products method (11) to perform a differential expression analysis across user-selected gene signatures. The method is a non-parametric statistic that detects items that are consistently highly ranked in a number of lists, for example genes that are consistently found among the most strongly over-expressed and under-expressed in a number of replicates. It has the advantage that it overcomes the heterogeneity among multiple datasets and does not require normalization.
In MARQ, users can select a set of signatures and the application creates a rank matrix with common genes in the different signatures. This matrix is analyzed using Rank products to find genes that are over-expressed and under-expressed, considering the different signatures as biological replicates in the differential expression analysis. This approach can provide useful information about common biomarkers across different experimental conditions.
MARQ APPLICATION
To run an analysis, users must input the query lists of genes and select the organism and the type of analysis (cross-platform or single platform). The gene Id format depends on the type of analysis; for platform specific queries users must use platform specific probe ids (e.g. Affymetrix probe Ids), but for cross-platform queries, several Id formats are supported, including Entrez, Ensemble, Gene symbols, most platform probe Id formats, and most of the Id formats listed in BioMart. Single platform analyses have a higher coverage on the number of probes but are limited to signatures from the same platform, as opposed to cross-platform analyses, which allow users to query all signatures in the database from a given organism. A single list can also be used (either considering it over- or under-expressed genes); this may prove interesting in some situations, for example, to explore what GEO signatures significantly modulate genes from a biological pathway.
After a job is launched, the user is redirected to a job status page that automatically reloads reporting progress status, until the job is finished. Once the analysis is completed the results page is loaded; it includes a table where rows represent signatures, sorted according to relevance to the query; signatures at the top are considered to be the most directly related to the query, while signatures at the bottom are the most inversely related. Signatures without enough replicates to perform the statistical test, and for which we sort genes by log-ratios instead of t-values, are marked with the ‘[ratio]’ keyword in their name.
The textual description of the signatures is linked to the ‘Word’ meta-information annotations described in a previous section, so clicking on a term highlights all signatures containing that word in their description. The ‘Annotations Menu’ can be used to explore other types of meta-information annotations such as GO terms.
The results page also includes a hit bar for each signature that shows the position of the query genes in it. The gene distribution can be carefully examined in the hit exploration page linked to each hit-bar that includes exact positions in the signature, gene names and links to the corresponding GEO profiles, if available.
Finally, users can select a list of signatures to include in the meta-analysis pipeline and the signature comparison page shows the genes that are commonly over-expressed and the genes that are commonly under-expressed across all the selected signatures.
Beyond the web tool, the core functionalities of MARQ are offered via a SOAP web service, which is used by the web portal application as the computational back-end. Using this technology, researchers can access MARQ functionalities programmatically to insert them in their data mining pipelines or in other bioinformatics systems; this can be done in a very straightforward manner. WSDL file and services description is available through the web portal help page. While the off-line processing involves using R, the on-line front end is coded entirely in Ruby, with some portions in in-lined C for efficiency. The complete source code for the applications back-end as well as for the web front end is available at http://github.com/mikisvaz, with instructions to configure and deploy a custom installation.
USE CASE: EXPLORING CELL WALL STRESS RESPONSES IN YEAST
To exemplify the potentials of the application, we have used MARQ to analyze the transcriptional program of the budding yeast S. cerevisiae to cell wall stress caused by zymolyase (12) which mainly affects the β-1,3glucan cell wall network. Yeast cells respond to cell wall damage by activating a transcriptional program that includes the up-regulation of genes mainly involved in cell wall remodeling, stress, metabolism and signaling (13). In addition MAPK kinase signaling pathways play important roles in the regulation of the transcriptional responses necessary for the adaptation of cells.
Analysis of the gene expression profile of a wild-type strain treated with zymolyase revealed that signatures derived from conditions such as exposure to Congo Red, long term exposure to DTT, hyperactivation of the cell wall integrity (CWI) pathway (GAL-PKC1-R398A), tunicamycin and hyper-osmotic conditions showed high positive scores with the zymolyase treatment. This is in agreement with the fact that all of these conditions affect cell wall integrity by different mechanisms. On the other hand, experiments related to hypo-osmotic conditions showed high negative scores.
The ‘compare signatures’ tool allowed us the identification of genes commonly over-expressed among the above mentioned cell wall stress conditions. Signatures related to zymolyase exposure, Dithiothrietol exposure and over-expression of elements of MAPK pathways (14) were used to determine common over-expressed genes. Using this approach we were able to identify the common cell wall compensatory ‘signature’ developed by the yeast in response to cell wall stress. Common genes include encoding cell wall-remodeling enzymes like CRH1 or BGL2, and other cell wall proteins such as YLR1194C, SED1, CWP1, PST1, PIR3 or YPS3, which corresponds with the fact that cell wall damage needs to be compensated by cell wall remodeling processes and cell wall-remodeling enzymes are the main components included in the common cell wall stress response (13) . Coupled with an increase in the chitin content, as part of this compensatory response, genes like GFA1, encoding for a protein involved in the biosynthesis of chitin, are included in the common over-expressed signature. In coherence with our previous findings (13), genes related to metabolism, stress and signaling were also identified. The last group is particularly interesting because two of the genes in this group include SLT2 and MLP1, both of them signaling components of the CWI pathway, the main pathway involved in regulating the transcriptional responses to cell wall stress.
Additional MARQ analysis of the yeast transcriptional response to zymolyase in a hog1 mutant allowed us to identify a crosstalk between the mating and HOG pathways. This result clearly illustrates the potential of this tool to identify novel connections between different signaling pathways. Full details of all these analysis are provided in the application web page.
In addition to the yeast analysis, another use case for a human melanoma study has been included in the Supplementary Data.
CONCLUSIONS
We present MARQ, a web-based application that enables users to query signatures derived from GEO, and retrieve experimental conditions that may induce similar or opposite gene expression programs to a given query. The system is provided in an easy-to-use and integrated environment that offers additional functionalities such as the possibility of performing gene expression meta-analysis to find common over- and under-expressed genes across different conditions, or connect signatures through common words or GO. We hope the application will prove useful to the research community.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish grants (BIO2007-67150-C03-02, S-Gen-0166/2006, TIN2005-5619, and PS-010000-2008-1). Funding for open access charge: European Union (project FP7-HEALTH-F4-2008-202047); Ministerio de Ciencia e Innovación (projects BIO2007-67150-C03-02, PS-010000-2008-1); Comunidad de Madrid (S-Gen-0166/2006).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 3.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 4.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newman JC, Weiner AM. L2L: a simple tool for discovering the hidden significance in microarray expression data. Genome Biol. 2005;6:R81. doi: 10.1186/gb-2005-6-9-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yi Y, Li C, Miller C, George AL., Jr Strategy for encoding and comparison of gene expression signatures. Genome Biol. 2007;8:R133. doi: 10.1186/gb-2007-8-7-r133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sean D, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 9.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995:289–300. [Google Scholar]
- 10.Efron B, Tibshirani R. On testing the significance of sets of genes. Ann. Appl. Stat. 2007;1:107–129. [Google Scholar]
- 11.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 12.Garcia R, Rodriguez-Pena JM, Bermejo C, Nombela C, Arroyo J. The high osmotic response and cell wall integrity pathways cooperate to regulate transcriptional responses to zymolyase-induced cell wall stress in Saccharomyces cerevisiae. J. Biol. Chem. 2009;284:10901–10911. doi: 10.1074/jbc.M808693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arroyo J, Bermejo C, Garcia R, Rodriguez-Pena J. Genomics in the detection of damage in microbial systems: cell wall stress in yeast. Clin. Microbiol. Infect. 2009;15:44–46. doi: 10.1111/j.1469-0691.2008.02676.x. [DOI] [PubMed] [Google Scholar]
- 14.Roberts CJ, Nelson B, Marton MJ, Stoughton R, Meyer MR, Bennett HA, He YD, Dai H, Walker WL, Hughes TR, et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.