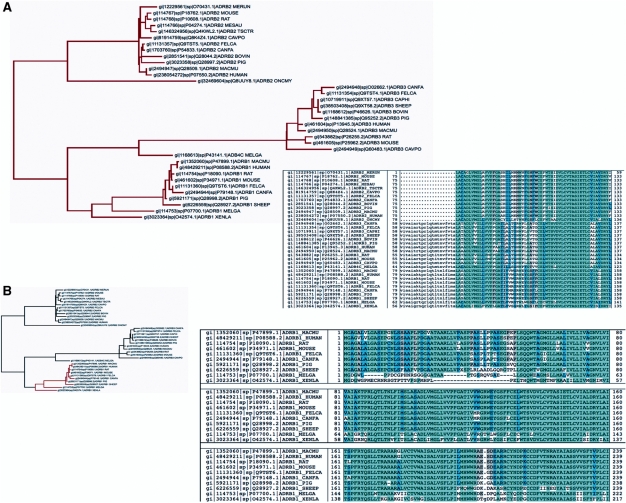
Figure 1.
Beta adrenergic receptors SATCHMO-JS tree and MSA displayed using the PhyloScope viewer. The PhyloScope viewer allows users to select internal nodes of the tree for examination of the alignments at these nodes, which may reflect different levels of inferred structural similarity across homologs. Columns are colored according to conservation based on BLOSUM62 sum-of-pairs scores (light blue indicates the highest level of conservation, followed by dark blue, grey and uncolored). Clicking on a subtree node restricts the MSA displayed to the sequences descending from that node, and highlights the selected subtree. The SATCHMO algorithm attempts to determine which columns are part of the conserved core structure across all sequences that descend from a node, resulting in some residues being displayed in lowercase (indicating that they are inserted relative to the consensus) at nodes higher in the tree (toward the root) but in uppercase at subtrees nearer the leaves. (A) The SATCHMO-JS tree and MSA corresponding to the root of the tree, where all sequences are selected. The first ∼70 residues of most sequences display in lowercase (indicating insertions relative to the consensus structure) reflecting structural variability over the dataset as a whole in this region. (Coincidentally, the region identified by SATCHMO as conserved across the dataset corresponds to the PFAM 7TM_1 HMM, which matches this region.) (B) The ADRB1 subtree (corresponding to orthologous Beta-1 adrenergic receptors from different species) has been selected by clicking the subtree node. This results in coloring the selected subtree red and displaying the MSA corresponding to sequences descending from that node. Note that many residues that displayed in lowercase in the SATCHMO root-level MSA are now displayed in uppercase, indicating that they are predicted by SATCHMO to be part of the conserve core structure for Beta-1 adrenergic receptors. Examining this subtree MSA shows that ADRB1_XENLA (from Xenopus laevis, African clawed frog) and ADRB1_MEGLA (from Meleagris gallopavo, Common turkey) diverge from mammalian orthologs at the N-terminus.