Table 3.
Validation for detection of specificity sites by SH and mR scored as area under curve (AUC) for the PR plots versus gold-standard specificity sites in the 22 data sets, 7 sets as defined in Table 2 and 15 sets obtained from Chakrabarti and Panchenko (15)
| Dataset | cbm9 | cd00 | cd00 | cd00 | cd00 | cd00 | cd00 | cd00 | CN- | GPCR | GPCR | GST | IDH/ | LacI | MDH/ | AQP/ | nucl | rab | ras/ | ricin | serine | Smad | Aver |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 120 | 264 | 333 | 363 | 365 | 423 | 985 | myc | 190 | IMDH | LDH | GLP | cycl.a | 5/6 | ral | Wt'd | ||||||||
| # positives | 7 | 3 | 3 | 12 | 6 | 10 | 4 | 3 | 11 | 21 | 21 | 9 | 14 | 28 | 1 | 23 | 2 | 28 | 12 | 21 | 2 | 29 | |
| mR | 0.161 | 0.058 | 0.006 | 0.301 | 0.010 | 0.055 | 0.204 | 0.329 | 0.037 | 0.246 | 0.347 | 0.156 | 0.050 | 0.266 | 0.063 | 0.213 | 0.417 | 0.540 | 0.666 | 0.186 | 0.078 | 0.719 | 0.310 |
mR Z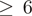
|
0.161 | 0.058 | 0.006 | 0.301 | 0.010 | 0.055 | 0.204 | 0.329 | 0.037 | 0.252 | 0.347 | 0.156 | 0.050 | 0.282 | 0.063 | 0.216 | 0.417 | 0.539 | 0.666 | 0.186 | 0.078 | 0.721 | 0.312 |
| SH. | 0.074 | 0.054 | 0.003 | 0.287 | 0.008 | 0.119 | 0.080 | 0.198 | 0.067 | 0.486 | 0.489 | 0.242 | 0.048 | 0.124 | 0.125 | 0.249 | 0.413 | 0.602 | 0.540 | 0.194 | 0.261 | 0.713 | 0.330 |
SH Z
|
0.074 | 0.054 | 0.003 | 0.287 | 0.008 | 0.119 | 0.080 | 0.198 | 0.067 | 0.517 | 0.489 | 0.242 | 0.048 | 0.207 | 0.125 | 0.268 | 0.413 | 0.602 | 0.540 | 0.194 | 0.261 | 0.703 | 0.342 |
| ProteinKeys | 0.049 | 0.008 | 0.087 | 0.203 | 0.010 | 0.010 | 0.002 | 0.034 | 0.027 | 0.377 | 0.505 | 0.483 | 0.065 | 0.301 | 0.005 | 0.119 | 0.011 | 0.364 | 0.092 | 0.276 | 0.006 | 0.748 | 0.287 |
| PROUST-II | 0.349 | 0.079 | 0.012 | 0.055 | 0.011 | 0.016 | 0.049 | 0.058 | 0.122 | 0.308 | b | 0.446 | 0.089 | 0.111 | 0.015 | 0.187 | 0.305 | 0.455 | 0.378 | 0.256 | 0.750 | 0.723 | 0.258 |
| SDPpred v.2 | 0.122 | 0.126 | 0.017 | 0.376 | 0.012 | 0.126 | 0.234 | 0.509 | 0.162 | 0.508 | 0.508 | 0.615 | 0.196 | 0.146 | 0.250 | 0.242 | 0.413 | 0.416 | 0.357 | 0.201 | 0.542 | 0.522 | 0.333 |
| Xdet | 0.352 | 0.106 | 0.080 | 0.366 | 0.011 | 0.103 | 0.196 | 0.387 | 0.086 | 0.125 | b | 0.117 | 0.100 | 0.190 | 0.033 | 0.169 | 0.054 | 0.350 | 0.398 | 0.173 | 0.105 | 0.688 | 0.234 |
| Xdet supc | 0.209 | 0.106 | 0.019 | 0.346 | 0.012 | 0.189 | 0.171 | 0.534 | 0.101 | 0.275 | b | 0.402 | 0.129 | 0.207 | 0.250 | 0.208 | 0.292 | 0.346 | 0.545 | 0.193 | 0.750 | 0.677 | 0.279 |
| Average | 0.172 | 0.072 | 0.026 | 0.280 | 0.010 | 0.088 | 0.136 | 0.286 | 0.078 | 0.344 | 0.448 | 0.318 | 0.086 | 0.204 | 0.103 | 0.208 | 0.304 | 0.468 | 0.465 | 0.206 | 0.314 | 0.691 | 0.298 |
aNucleotidyl cyclase.
bThe GPCR data set is above the maximum of 1000 sequences for these methods.
cSupervised by using subgroupings.
A higher AUC corresponds to better performance. For comparison, predictions by ProteinKeys, PROUST-II, SDPpred v.2 and Xdet are also shown. Best-scoring methods for each data set are in bold. The final column list the average AUCs per method weighted by number of positives, and the bottom row the averages per data set.
