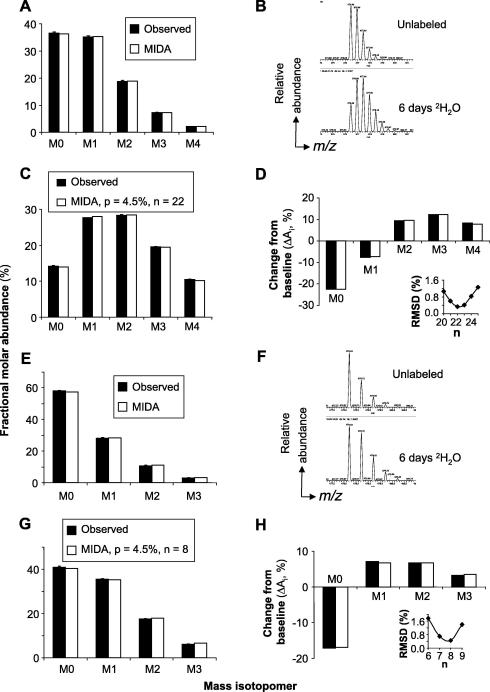
Fig. 3.
Mass isotopomer distributions of HLA-DR4-derived tryptic peptides. (A–D) Analysis of DRα (51–67) peptide. (A) MIDA model and experimental quantification (means ± SDs of N = 6 replicate injections) of mass isotopomer distributions of the unlabeled DRα peptide. (B) Mass spectrum of the DRα peptide obtained before and after 6 days of culturing Priess cells in 4.5% 2H2O. (C) MIDA model and experimental quantification (means ± SDs, N = 5) of mass isotopomer distributions after 2H2O labeling. The model assumes that n = 22 labeling sites are accessible to label without dilution of the precursor pool enrichment (p) relative to the 2H2O enrichment in medium; the value of n was adjusted for optimal fit to the data. (D) The difference between the mass isotopomer distributions of the fully labeled and unlabeled DRα peptide is shown. The inset shows the RMSD of the experimental data from a series of models in which p was kept at 4.5% and n was varied. The lowest RMSD value was obtained at n = 22. (E–H) The same analysis was applied to the DR0401β (73–80) peptide: (E) unlabeled mass isotopomer distributions (means ± SDs, N = 5 replicates); (F) mass spectra of unlabeled versus extensively labeled peptide; (G) mass isotopomer distributions of labeled peptide (means ± SDs of N = 5 replicate analyses, RMSD = 0.45% vs. model with n = 8 labeling sites and p = 4.5%); (H) difference analysis and dependence of model fit on n (inset).